In application scenarios such as species identification, evolutionary research and ecological monitoring, the mitochondrial genome is known as the "super barcode" due to its inherent advantages including high resolution, stable genetic composition and an optimal evolutionary rate. It can not only accurately distinguish interspecific differences but also reflect the evolutionary context of taxa, thus providing critical technical support for the study of complex biological issues. Based on its independently developed TargetSeq® liquid-phase hybridization capture and sequencing technology, iGeneTech has created a multi-species mitochondrial capture solution that covers the mitochondrial genomes of pan-taxa species including chordates, arthropods, green plants and fungi. Customers can select products according to the taxonomic classification of their samples, and the solution is applicable to research directions such as species identification (for scientific research, judicial and customs purposes), evolutionary studies, population genetics and ancient DNA capture.
Focusing on the Challenges of Species Identification, Mitochondrial Capture Technology Highlights Distinct Advantages
Traditional morphological identification and DNA barcoding can meet the needs of routine species recognition, yet they fail to accurately identify problematic species with inconspicuous morphological characteristics or ambiguous sequence alignments. Although whole-genome sequencing yields accurate results, it has limitations such as massive data volume, high cost, long turnaround time and high technical threshold, making it unsuitable for large-scale scientific research scenarios.
Multi-species Mitochondrial Capture: Balancing Precision and Practicality
1. High Coverage
Designed based on more than 18,000 full-length mitochondrial sequences from the NCBI database, the product achieves a coverage rate of up to 99.9%, ensuring no omissions in target regions.
2. High Enrichment Efficiency
The maximum number of probes in a single panel exceeds 1.49 million. The high-density design not only improves the specificity of targeted binding but also enhances compatibility with different samples of the same taxon.
3. High Cost-Effectiveness
The estimated data volume is only 0.5-1 Gb, a reduction of more than 90% compared with whole-genome sequencing. The solution also supports multiplex capture protocols, effectively reducing experimental costs and analysis cycles.
4. Broad Applicability
Covering 7 major taxa including chordates, arthropods, green plants and fungi, it is adaptable to a variety of research directions.
5. Strong Compatibility
Suitable for fresh samples as well as complex samples such as ancient DNA, degraded DNA and environmental DNA (eDNA), breaking through the limitations of sample quality.
Pan-taxa Product Matrix: Precisely Matching Diverse Research Needs
Product Name | Main Covered Taxa | Core Parameters |
Chordate Mitochondrial Capture | Vertebrates such as mammals, birds and fishes, as well as cephalochordates and urochordates | 7,865 mitochondrial genomes, 99.6% coverage, over 890,000 probes |
Arthropod Mitochondrial Capture | Insecta, Arachnida, Crustacea and other taxa such as ants, butterflies, spiders and crabs | 6,083 mitochondrial genomes, 98.3% coverage, over 840,000 probes |
Mollusk Mitochondrial Capture | Bivalvia, Cephalopoda, Gastropoda and other taxa such as freshwater mussels, oysters, squids and snails | 870 mitochondrial genomes, 99.6% coverage |
Green Plant Mitochondrial Capture | Chlorophyta, Charophyta, as well as bryophytes, pteridophytes and seed plants | 688 mitochondrial genomes, 99.9% coverage |
Fungal Mitochondrial Capture | Various fungal taxa | 891 mitochondrial genomes, 97.7% coverage |
Mitochondrial Capture for Other Taxa | Protozoa such as Apicomplexa and Ciliophora, as well as invertebrates | 1,817 mitochondrial genomes, 97.5% coverage, over 290,000 probes |
Mitochondrial Capture for 242 Mammalian Species | 242 specific mammalian species | 242 mitochondrial genomes, 100% coverage |
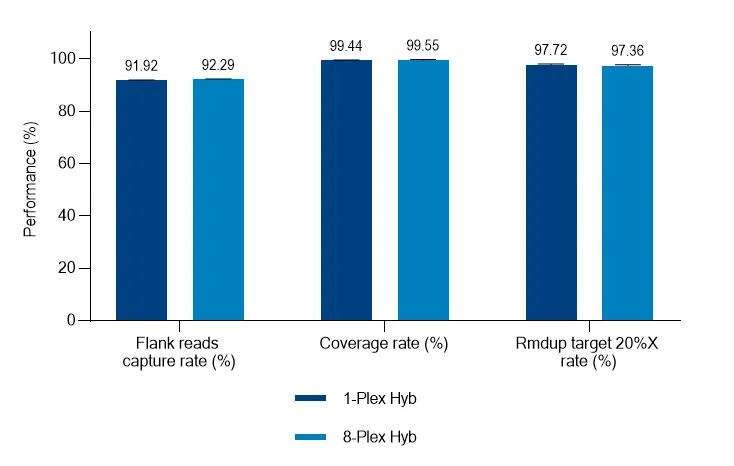
Real Test Data Demonstration
Multiplex capture was performed on 6 species including chickens, dogs and fishes with the Chordate Mitochondrial Capture Panel, and all tested samples were accurately identified to the species level.
Known Information | Mitochondrial Identification Result | Sequence Name | Coverage rate (%) | Target mean depth (×) |
Chicken | NC_040902 | Red Junglefowl (Southern Yunnan subspecies) | 100 | 2485 |
Dog | NC_008092 | Gray Wolf (ancestor of domestic dogs) | 100 | 71036 |
Mouse | NC_006914 | Mouse | 100 | 51588 |
Fish | NC_018791 | Nanhai Beltfish | 97.9 | 26230 |
Fish | NC_041308 | Bohadschia callista | 99.9 | 3353 |
Fish | NC_001960 | Atlantic Salmon / Salmon | 100 | 25685 |
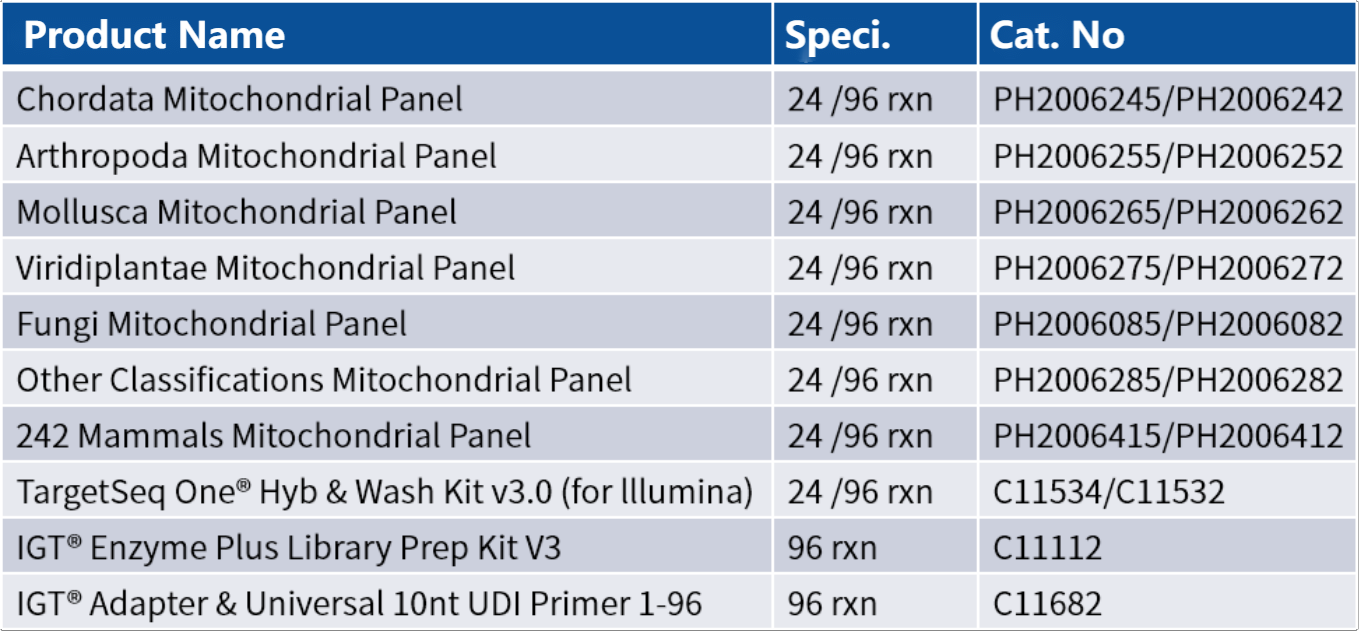
Product Info

 CN
CN