iGeneTech New Product Release
As a crucial model organism for human disease research and drug development, the mouse whole exome product plays a vital role in life science research. By sequencing targeted exonic regions, mouse whole exome products efficiently and cost-effectively reveal variant information in the genome's coding regions, providing powerful tools for disease mechanism research, drug development, and gene function exploration.
Relying on the self-developed IGT® Oligo Pools Synthesis Platform and TargetSeq® Technology Platform, iGeneTech has developed mouse whole exome products.
iGeneTech's Mouse Whole Exome Panel is designed based on the 38.429 Mb CDS region of the mm39 reference genome, with 523,352 unique probes, enabling efficient and uniform capture of the mouse CDS region.
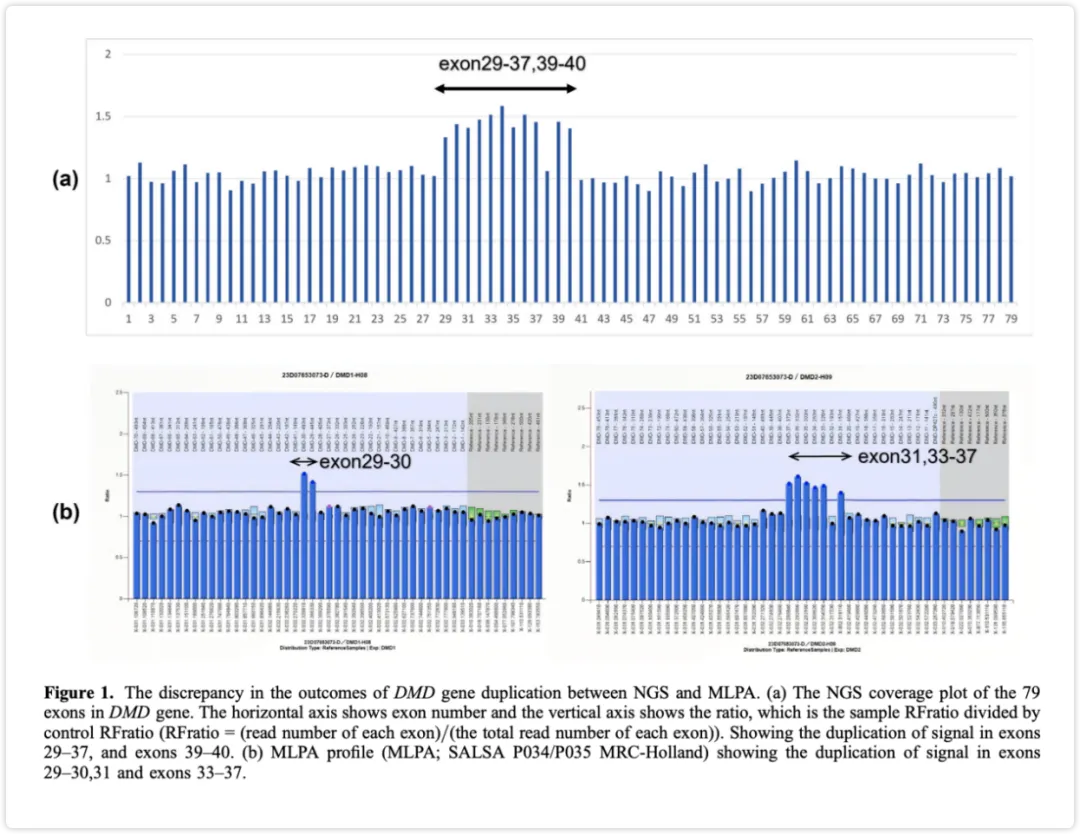
Compared with Whole Genome Sequencing (WGS), whole exome sequencing (WES) focuses on exonic regions, offering lower sequencing costs and smaller data volumes. The mouse genome is approximately 2.77 Gb—for example, 100-fold WGS requires at least 300 Gb of data. iGeneTech's actual test data shows that 8 Gb achieves a depth of over 100-fold, demonstrating excellent performance metrics.
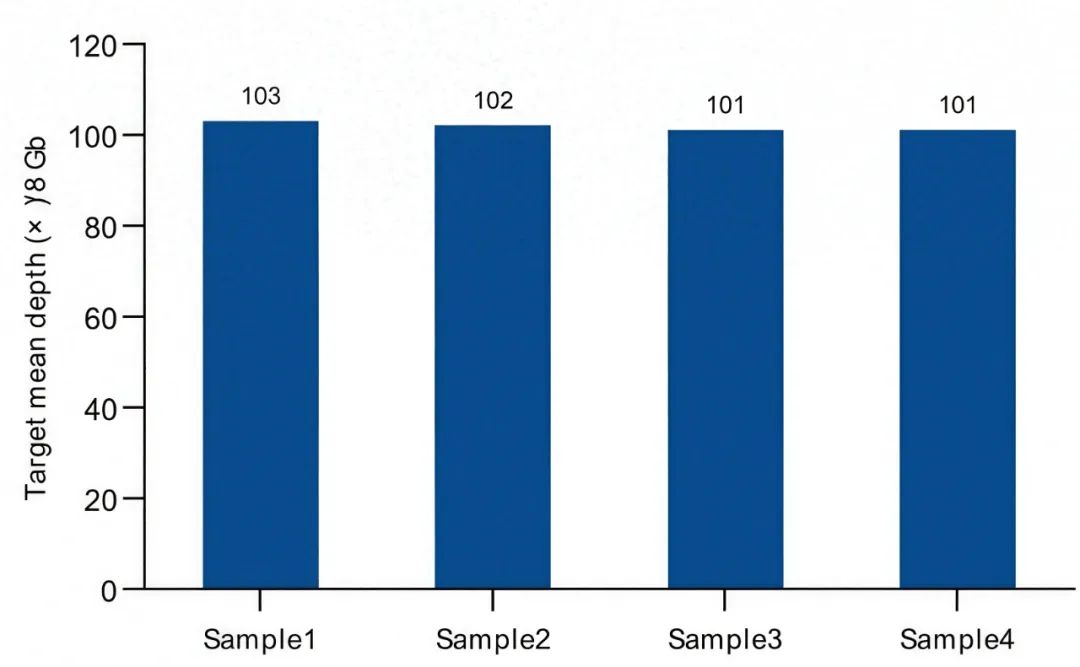
The Mouse Whole Exome Panel, combined with the TargetSeqOne® hybridization capture system, enables efficient enrichment of target regions with a capture efficiency of over 75%. Meanwhile, this product demonstrates high uniformity: the 20%X uniformity exceeds 98%. With the same data volume, it effectively improves the coverage and accuracy of exome detection.
As of now, igenetech has launched over ten whole exome products for animals and plants. More whole exome products are under continuous development. All products support full customization and semi-customization.
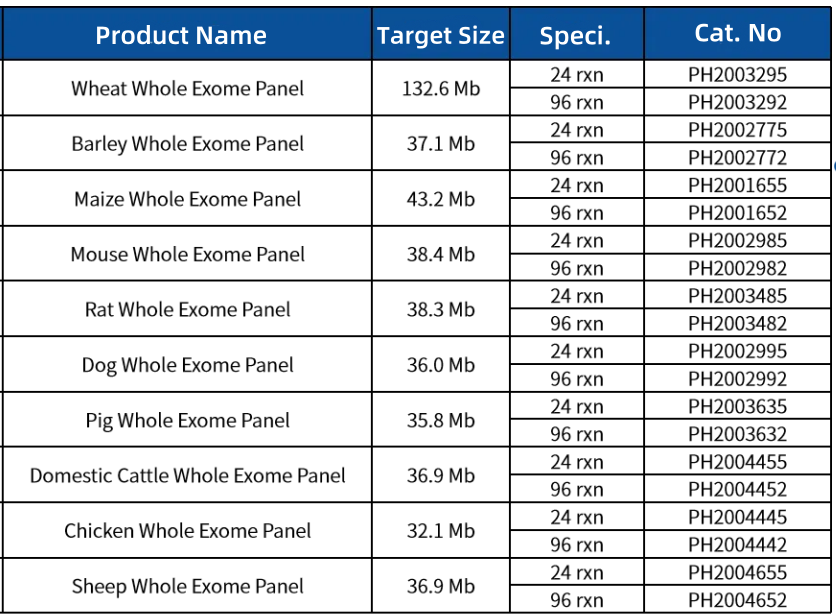
Order Information

 CN
CN