ABX Guide-Diagnosis and Treatment of Infectious Diseases is a practical reference book compiled by experts from the Johns Hopkins University School of Medicine and translated by Professor Ma Xiaojun's team from Peking Union Medical College Hospital.
The book systematically elaborates on the diagnostic criteria for infectious diseases, pathogen characteristics, treatment principles, and the application of antibacterial agents. It covers infections caused by various pathogens such as bacteria, viruses, fungi, and parasites, providing clinicians with reference information on antibacterial agents, infectious diseases, and common pathogens.
It is a quick-reference guide for medication and a learning tool suitable for clinicians in all clinical departments.

Based on the diagnosis and treatment concepts of this book, iGeneTech officially launches the ABX Guidelines Pathogen Precision Capture Probe Solution.
This solution focuses on 78 bacterial species, 12 fungal species, and 26 viral species covered in the guidelines. It independently develops highly specific capture probes for each type of pathogen, with a coverage rate exceeding 99% for all.
Table 1. Bacteria Involved in Diagnosis and Treatment of ABX Infectious Diseases |
Acinetobacter baumannii | Actinomycete | Aeromonas | Bacillus | Bacteroides fragilis | Bacteroides |
Bartonella | Bordetella | Borrelia | Brucella | Burkholderia cepacia complex | Burkholderia mallei |
Campylobacter | Campylobacter jejuni | Capnocytophaga canimorsus | Chlamydia trachomatis | Chlamydia pneumoniae | Chlamydia psittaci |
Citrobacter | Clostridium botulinum | Clostridium difficile | Clostridium | Clostridium tetani | Corynebacterium diphtheriae |
Coxiella burnetii | Ehrlichia | Eikenella corrodens | Enterobacter | Enterococcus | Erysipelothrix rhusiopathiae |
Escherichia coli | Francisella tularensis | Haemophilus ducreyi | Haemophilus influenzae | Moraxella catarrhalis | Klebsiella |
Lactobacillus | Legionella | Leptospira | Listeria monocytogenes | Morganella | Mycobacterium abscessus |
Mycobacterium avium Complex | Mycobacterium chelonae | Mycobacterium fortuitum | Mycobacterium kansasii | Mycobacterium leprae | Mycobacterium marinum |
Mycoplasma pneumoniae | Neisseria gonorrhoeae | Neisseria meningitidis | Nocardia | Pasteurella multocida | Peptostreptococcus |
Plesiomonas | Propionibacterium | Proteus | Providencia | Pseudomonas aeruginosa | Rhodococcus equi |
Rickettsia rickettsii | Rickettsia | Salmonella | Shigella dysenteriae | Shigella | Staphylococcus |
Staphylococcus aureus | Stenotrophomonas maltophilia | Streptococcus moniliformis | Streptococcus pneumoniae | Streptococcus pyogenes | Streptococcus |
Treponema pallidum | Tropheryma whipplei | Vibrio cholerae | Vibrio | Yersinia pestis | Yersinia |
Table 2. Fungi Involved in Diagnosis and Treatment of ABX Infectious Diseases |
Aspergillus | Blastomyces dermatitidis | Candida albicans | Candida | Coccidioides immitis | Cryptococcus neoformans |
Dermatophyte | Fusarium | Histoplasma capsulatum | Scedosporium boydii | Sporothrix schenckii |
|
Table 3. Viruses Involved in Diagnosis and Treatment of ABX Infectious Diseases |
Mastadenovirus | Cytomegalovirus | Enterovirus | Epstein-Barr virus |
Hantaviridae | Hepatitis A virus | Hepatitis B virus | Hepacivirus hominis |
Herpes simplex virus | human gammaherpesvirus 8 | Hepatitis D virus | human Papillomavirus |
JC virus × BK virus | Measles morbillivirus | Molluscum contagiosum virus | Mumps orthorubulavirus |
Norovirus | Respirovirus | Human parvovirus B19 | Lyssavirus |
Human respiratory syncytial virus | Rhinovirus | Rubella virus | Zoster virus |
West Nile virus | Human T- lymphotropic virus (HTLV) |
|
|
Research Example 1: Analysis of HPV Integration Mechanism
Persistent HPV infection is recognized as the primary risk factor for cervical cancer [1]. The team from the Department of Obstetrics and Gynecology, Tongji Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology, performed whole-genome sequencing (WGS) with 55–65× coverage in SiHa and HeLa cells using the PacBio long-read sequencing platform. Through comprehensive analysis of sequence data, the study revealed the complexity of HPV integration. The accuracy of this research is comparable to that of the targeted capture next-generation sequencing (NGS) method [2].
iGeneTech provided customized liquid-phase hybridization capture technology and corresponding target enrichment solutions for this study.

Figure 1 HPV Integration Mechanisms in SiHa and HeLa Cells
Research Example 2: A Novel Strategy for HBV Integration Detection
The Molecular Laboratory of Infectious Diseases, Chongqing Medical University, developed an improved strategy based on DNA probe capture and next-generation sequencing (NGS) for HBV integration site detection. This study confirmed that using this optimized strategy, HBV DNA integration can be detected in the plasma cell-free DNA (cfDNA) of HBV-infected patients, including those with chronic hepatitis B (CHB), liver cirrhosis (LC), or hepatocellular carcinoma (HCC) [3].
iGeneTech provided multiple HBV genome hybridization capture technologies and corresponding library construction solutions for this research.
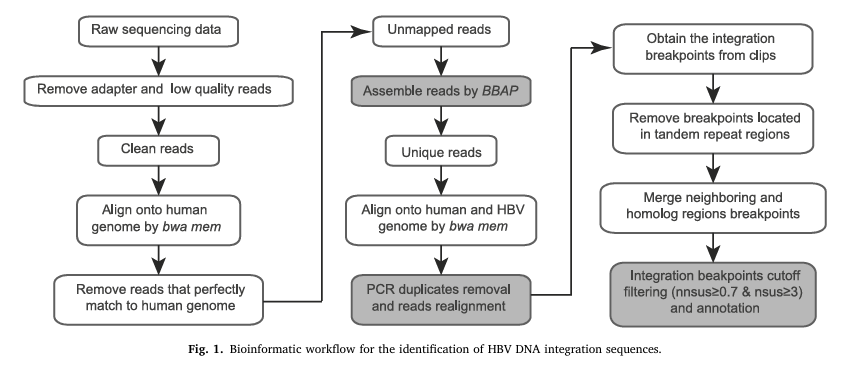
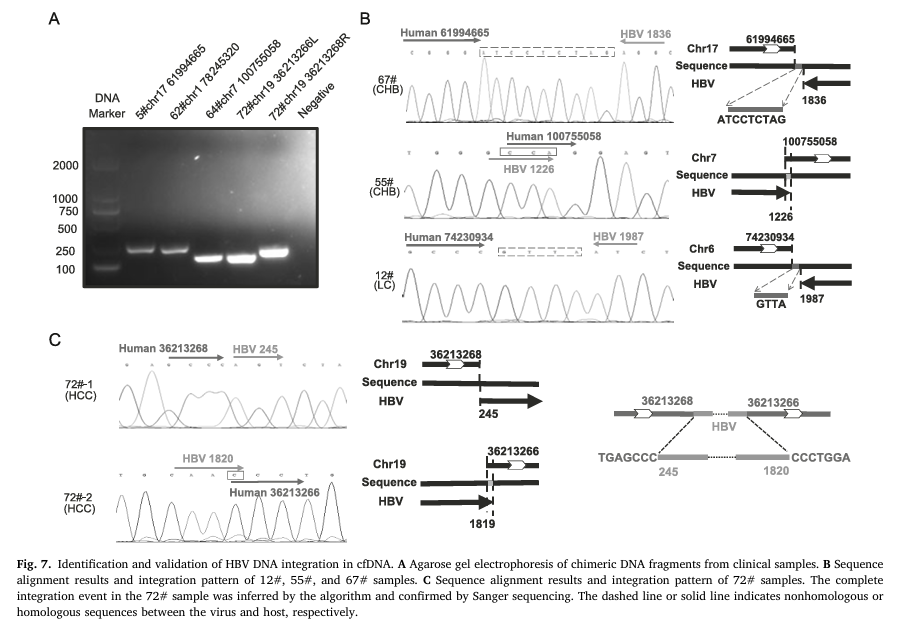
Figure 2 Bioinformatics Analysis Pipeline, Identification, and Validation of HBV DNA Integration in cfDNA
Probe Design Scheme
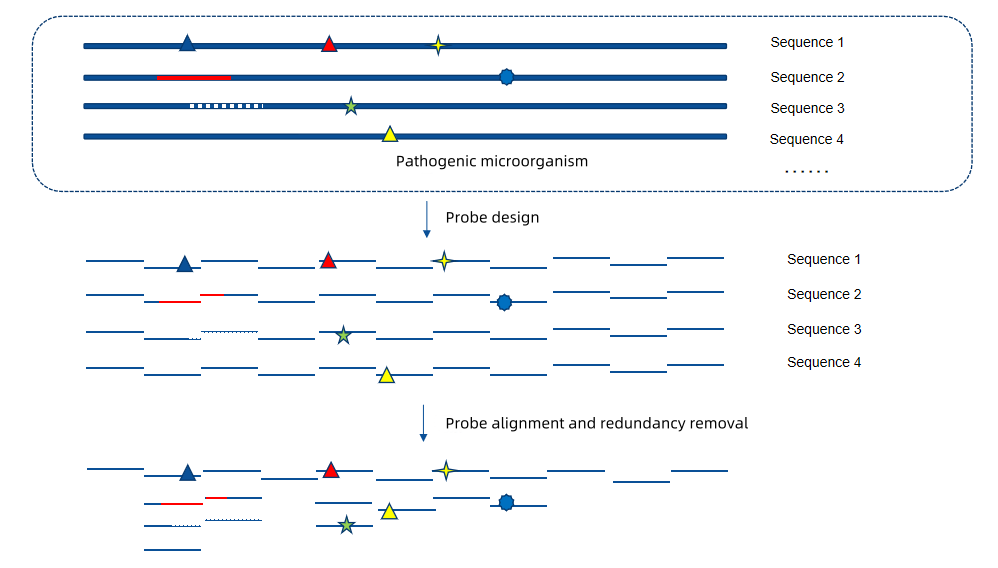
Figure 3 Probe Design Scheme
This scheme is based on the proprietary TargetSeq® liquid-phase probe hybridization capture technology. During the probe design phase, sequences of all strains in the NCBI database are used as references to design diverse and specific probes. These probes can cover multiple variants of pathogenic microorganisms, are compatible with emerging mutations of microorganisms, and avoid missed detections.
Technical Features
Ultra-broad spectrum coverage, no fear of sequence variations
Probe pools are designed based on sequences of multiple strains in the NCBI database, covering various pathogen subtypes. They effectively capture different variants and address regional differences as well as emerging mutations.
High specificity, rejecting cross-reactions
In complex clinical samples (such as environmental samples and sewage samples), probes undergo rigorous bioinformatics screening and wet experiment validation. They can accurately distinguish homologous sequences and effectively avoid interference from human background nucleic acids and symbiotic flora.
Excellent sensitivity, capturing weak signals
Targeting the difficulty of low-load infections, probe densification optimization achieves efficient binding and enrichment of target sequences. Even trace amounts of pathogen nucleic acids can be stably captured, providing guarantee for high-sensitivity detection.
Performance
Capture performance tests have been conducted on various pathogens, with specific data as follows:
1. Mycoplasma pneumoniae
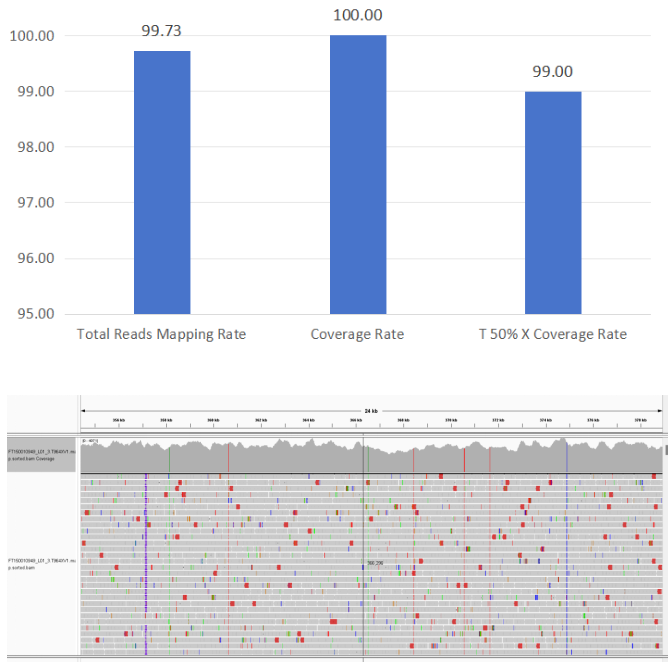
Figure 4 Full-Length Capture Performance of Mycoplasma pneumoniae
2. Epstein-Barr virus (EBV)
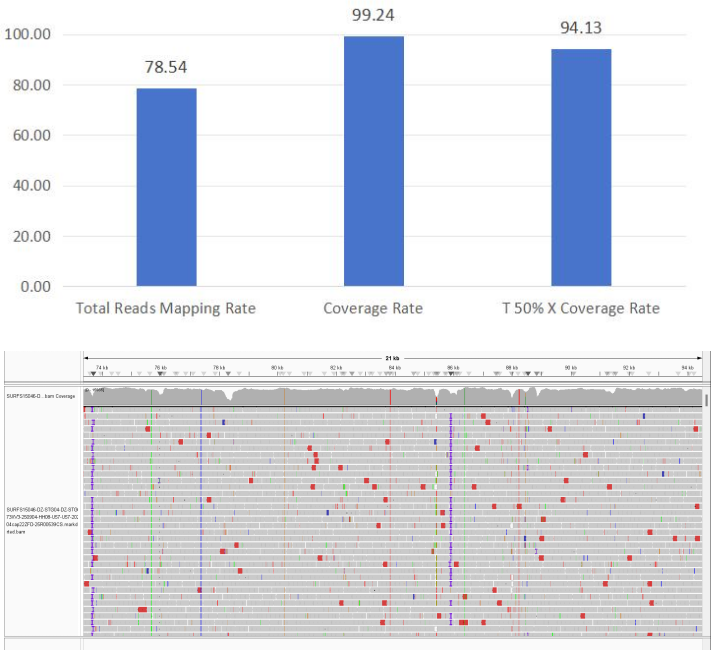
Figure 5 Full-Length Capture Performance of Epstein-Barr Virus (EBV)
3. Vibrio cholerae
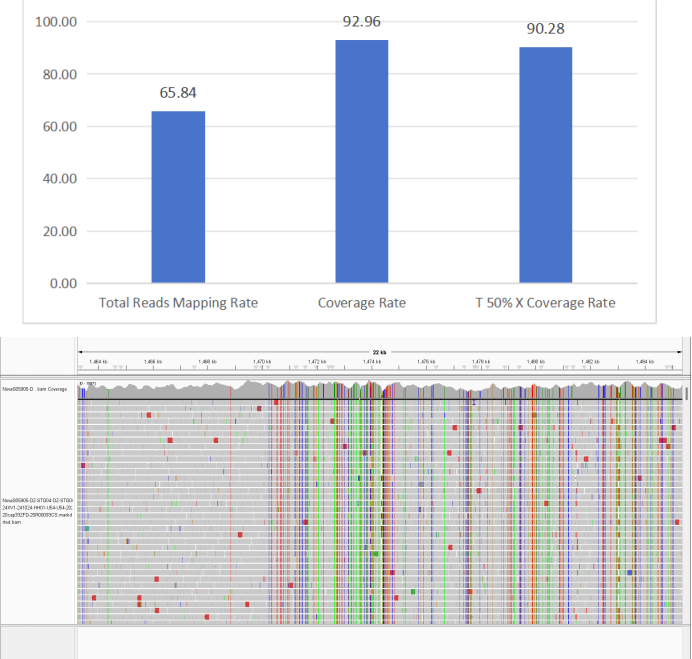
Figure 6 Full-Length Capture Performance of Vibrio cholerae
4. Human Papillomavirus (HPV)
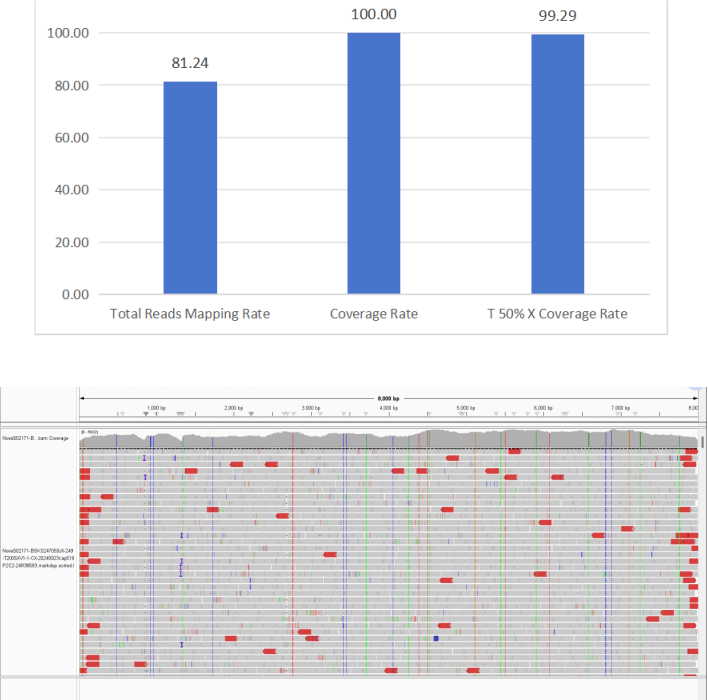
Figure 7 Full-Length Capture Performance of Human Papillomavirus (HPV)
5. Norovirus
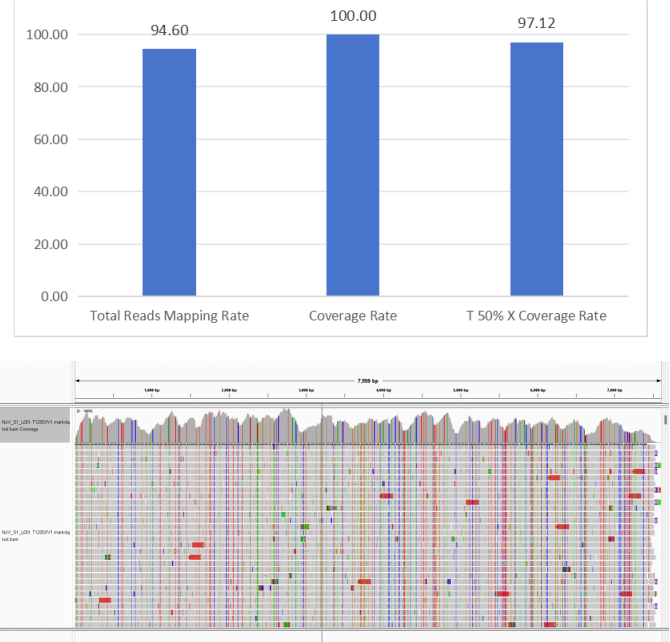
Figure 7 Full-Length Capture Performance of Norovirus
References:
[1] Bonin CM, Padovani CTJ, da Costa IP, Ávila LS, Ferreira AMT, Fernandes CES, Dos Santos AR, Tozetti IA. Detection of regulatory T cell phenotypic markers and cytokines in patients with human papillomavirus infection. J Med Virol. 2019 Feb;91(2):317-325. doi: 10.1002/jmv.25312. Epub 2018 Sep 24. PMID: 30192406.
[2] Wang Z, Liu C, Liu W, Lv X, Hu T, Yang F, Yang W, He L, Huang X. Long-read sequencing reveals the structural complexity of genomic integration of HPV DNA in cervical cancer cell lines. BMC Genomics. 2024 Feb 20;25(1):198. doi: 10.1186/s12864-024-10101-y. PMID: 38378450; PMCID: PMC10877919.
[3] Yang Z, Zeng J, Chen Y, Wang M, Luo H, Huang AL, Deng H, Hu Y. Detection of HBV DNA integration in plasma cell-free DNA of different HBV diseases utilizing DNA capture strategy. Virol Sin. 2024 Aug;39(4):655-666. doi: 10.1016/j.virs.2024.06.003. Epub 2024 Jun 7. PMID: 38852920; PMCID: PMC11401475.

 CN
CN










