In the clinical diagnosis of genetic diseases, sufficient coverage, stable depth, and successful capture of key loci often determine the success of a report.
Based on the globally authoritative OMIM database (Online Mendelian Inheritance in Man), iGeneTech further integrates high-evidence-level loci from ClinVar data (ClinVar Pathogenic/Likely Pathogenic variants) and the full mitochondrial DNA (mtDNA) genome sequence. This has led to the creation of the new medical whole-exome product AlExome® Human Medical Exome, upgrading it from a traditional whole-exome to a professional solution that is more focused on clinical value, delves deeper into pathogenic regions, and simultaneously covers both nuclear and mitochondrial genes.
Core Highlight 1: 5019 Clinical Core Genes
Backed by OMIM AuthorityStarting from OMIM, we have selected 5019 high medical value genes closely associated with human monogenic diseases. These genes fully cover 14 major disease categories, including the nervous system, cardiovascular system, metabolic and endocrine system, hematological system, immune system, and cancer susceptibility syndromes, ensuring the detection range is neither overly broad nor incomplete.
Table 1: Partial Disease Categories and Common Representative Genes from the OMIM Database
Type | Common associated genes |
Genetic diseases of the nervous system | SMN1、HTT、SCN1A |
Genetic diseases of the cardiovascular system | KCNQ1、MYH7、FBN1 |
Genetic and endocrine metabolic diseases | PAH、LDLR、G6PC |
Hematological and immunological genetic diseases | HBB、IL2RG |
Cancer susceptibility syndromes | BRCA1、MLH1、TP53 |
Ocular genetic diseases | RHO、CEP290 |
Genetic hearing loss disorders | GJB2、MITF |
Genetic diseases of the skin and hair | TYR、KRT5 |
Core Highlight 2: Comprehensive Supplement of ClinVar Key Mutation Sites, Addressing the Pain Point of Interpreting Variants of Uncertain Significance (VUS)
AlExome® Human Medical Exome takes the gene CDS ±20bp as the target region, adds ClinVar P/LP mutation sites, and incorporates important genetic disease-related genes with special probe designs*. This significantly improves detection performance.

*Specially designed genes include SMN1, SMN2, thalassemia-related regions, DMD, CYP2D6, CYP21A2, PKD1, etc.
Core Highlight 3: Stable and High-Depth Sequencing of Full-Length Mitochondrial Genome, Simultaneous Monitoring of Two Systems
Mitochondrial diseases often present with multisystem involvement in clinical practice, and traditional nuclear gene testing usually fails to cover mtDNA variants.
We have included the entire mitochondrial genome in the capture range:
l 100% coverage
l One-time detection of mtDNA SNVs/InDels
l High-depth mitochondrial regions enabling heteroplasmy level analysis
l Combined interpretation of clinical phenotypes with nuclear gene results
Truly achieving "one run, dual-system analysis" for both nuclear genes and mitochondrial genes.
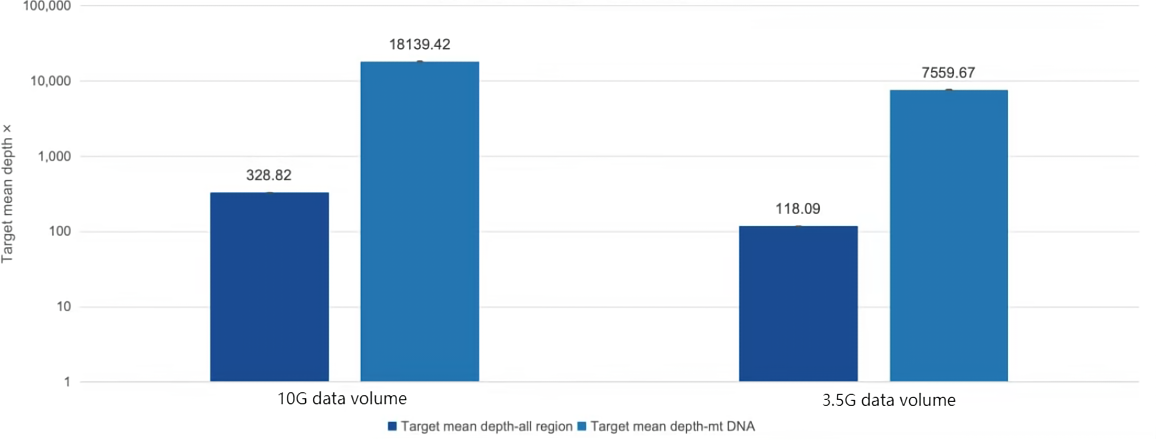
Figure 1. Excellent Mitochondrial Depth Ratio of AlExome® Human Medical Exome
200 ng of standard nucleic acid (Promega G3041) was used for enzymatic digestion library construction, followed by overnight hybridization capture with AlExome® Human Medical Exome probes. PE150 sequencing was performed on ZymoSurf SURFSeq 5000. Data was randomly sampled to 10 Gb and 3.5 Gb, and the average depth of the complete target region and full-length mitochondrial genome was calculated. The average mitochondrial depth can reach more than 50 times that of the target region.
Core Highlight 4: Lighter, Higher Coverage, and More Suitable for Clinical Applications
Compared with traditional research-oriented WES, AlExome® Human Medical Exome has a more focused target region. A data volume of 3.5 Gb can achieve a sequencing depth of over 100x, and 10 Gb can reach over 300x. The 30x coverage rate exceeds 99%, enabling effective identification of low-frequency heterozygous mutations and chimeric pathogenic mutations. It improves the detection confidence of SNPs/InDels, significantly reduces false negative and false positive rates, and enhances the credibility of reports.
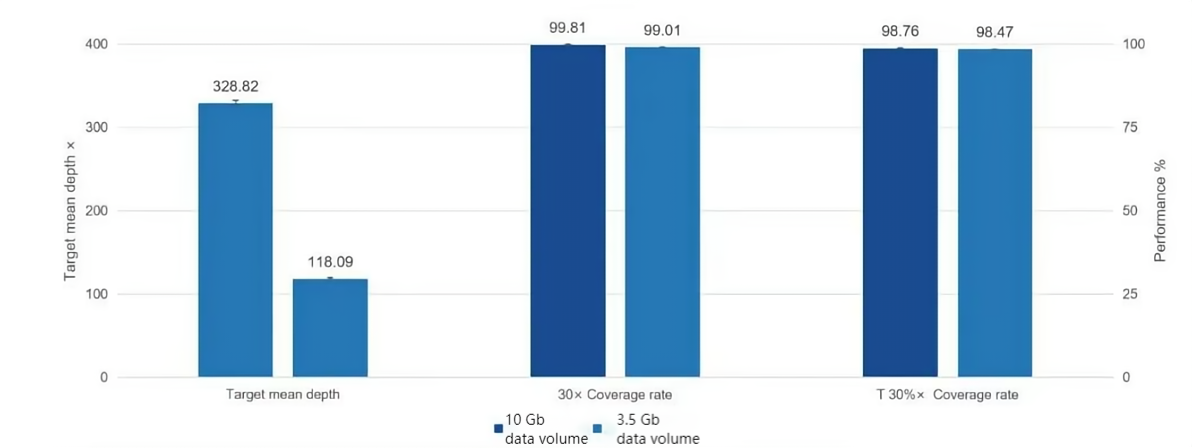
Figure 2. Excellent Coverage of AlExome® Human Medical Exome
200 ng of standard nucleic acid (Promega G3041) was used for enzymatic digestion library construction, followed by overnight hybridization capture with AlExome® Human Medical Exome probes. PE150 sequencing was performed on ZymoSurf SURFSeq 5000. Data was randomly sampled to 10 Gb and 3.5 Gb. At 3.5 Gb, the 30x coverage rate reaches 99% and the 30%x coverage rate reaches 98.47%; at 10 Gb, the 30x coverage rate reaches 99.81% and the 30%x coverage rate reaches 98.76%.
A medical whole-exome is more than just “testing”--it is a systematic upgrade centered on clinical diagnostic value.
We establish an accurate mapping between diseases and genes based on OMIM, carefully select and annotate evidence for each variant with reference to ClinVar, and integrate a new generation of capture and sequencing systems to ensure high coverage, high consistency, and reproducibility.
AlExome® Human Medical Exome is designed for real clinical scenarios—to provide doctors with more interpretable reports, genetic counselors with more evidence-based data, and help laboratories strike a balance between efficiency and accuracy. It is not a simple "upgraded test" but a redesigned whole-exome solution truly serving diagnosis and decision-making.
Product Info
Product Name | Speci. | Cat. No |
AlExome® Human MedicaI Exome | 24 / 96 rxn | PH2013005/PH2013002 |
TargetSeq One® Hyb & Wash Kit v3.0 with Eco Universal Blocking Oligo (for lllumina) | 24 / 96 rxn | C11614 /C11612 |
TargetSeq One® Hyb & Wash Kit v3.0 with Eco Universal Blocking Oligo (forMGI DI) | 24 / 96 rxn | C11624 /C11622 |
IGT® Enzyme Plus LibraryPrep Kit V3 | 96 rxn | C11112 |
IGT® Adapter & 10 nt UDlPrimer 1-768(for lllumina,plate) | 768 rxn | C11282 |

 CN
CN