As the proportion of genetic diseases in birth defects increases year by year[1], traditional prenatal and neonatal screening methods can hardly meet the need for accurate identification of rare and complex monogenic diseases. Whole Exome Sequencing (WES), a high-throughput sequencing technology covering all protein-coding regions, is gradually being applied in prenatal diagnosis and neonatal screening.
Studies have shown that WES has unique advantages in identifying de novo mutations and rare genetic diseases[2]. In addition, among neonates with atypical postnatal phenotypes or rapidly progressive conditions, WES helps shorten the time to diagnosis, enabling early diagnosis and treatment.
As an important supplement to traditional methods, WES is becoming a key tool for precision screening and intervention of hereditary diseases, providing more reliable molecular evidence for reproductive decision-making, prenatal management, and neonatal health protection[3].
iGeneTech focus on the probe capture technology field for a decade, committed to breaking international monopoly through underlying technological innovation. Recently, iGeneTech has grandly unveiled the AIExome® V5 family with three major matrices: Core Edition, Tumor Edition, and Inherit Edition. With continuous breakthroughs and innovations in performance and technology, it has broken the constraints of international brands and redefined the competitive landscape of the industry.
Core Features of the V5 Series: Multiple technical capabilities exceed industry standards
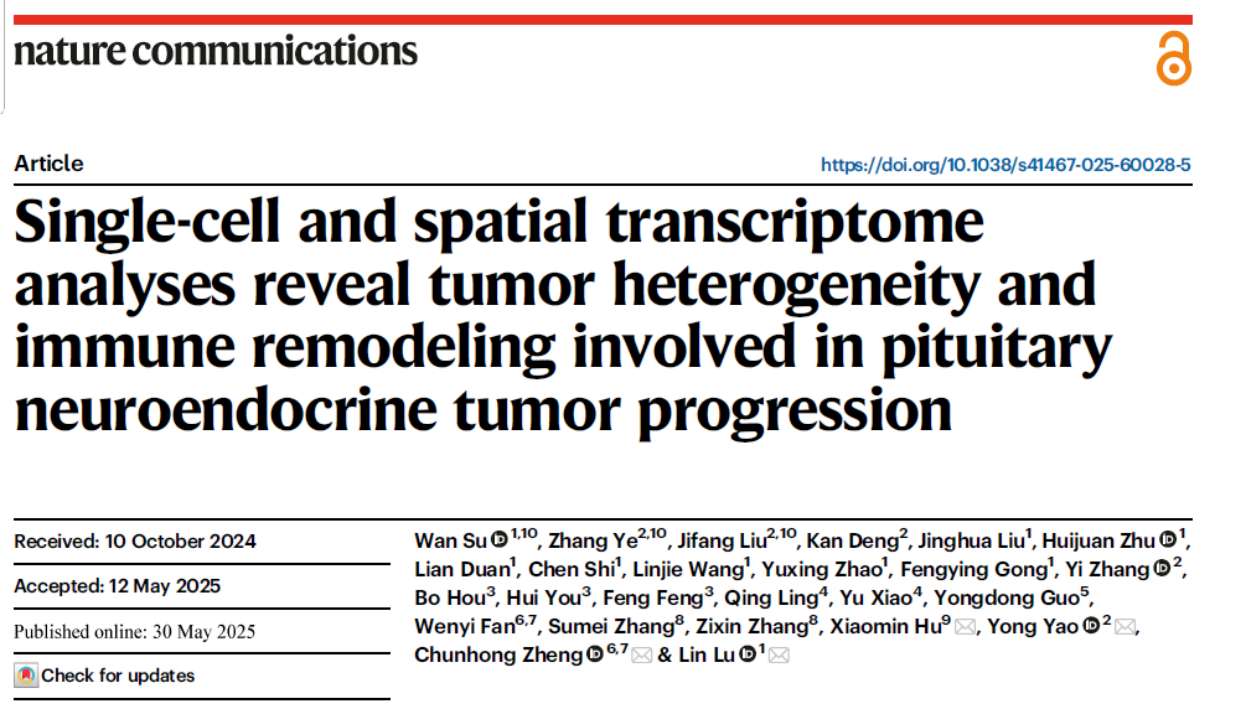
01 Outstanding Uniformity Performance
The V5 Core Edition features ultimate uniformity with a Fold 80 of 1.24. When combined with the 2-hour rapid hybridization capture protocol, this indicator remains stably controlled at 1.5, breaking the long-standing technical barriers monopolized by international brands and providing more reliable underlying data support for variant detection.
The data indicators are as follows:
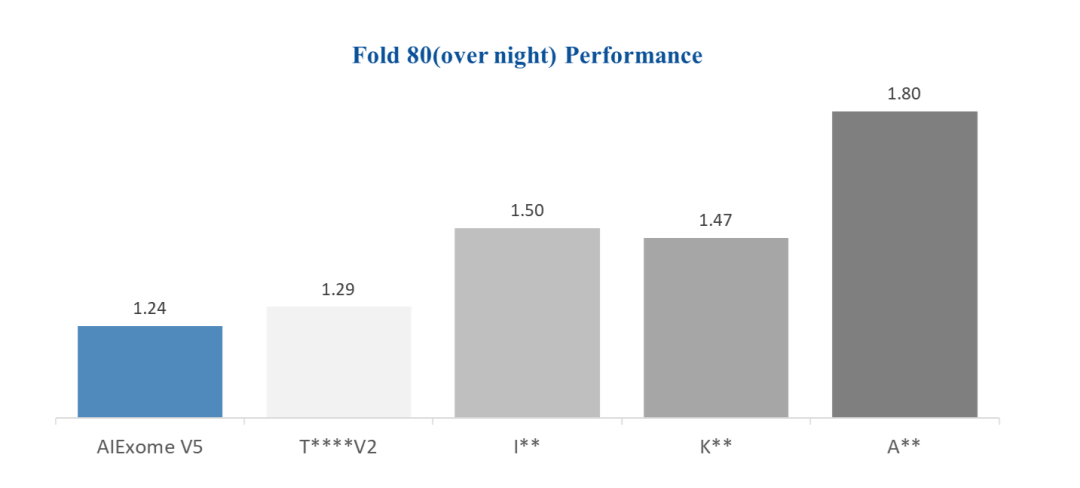
02 Genome-wide Precise Coverage
Through multi-layered tile probe layout and intelligent probe screening technology, the V5 probes balance coverage and uniformity. At an average depth of 100×, the 50× coverage rate reaches over 96%. With the same data volume, the rates of 20× and 30× coverage are higher, and there are fewer exons with poor coverage in OMIM genes, truly achieving "zero-blind-spot" gene scanning.
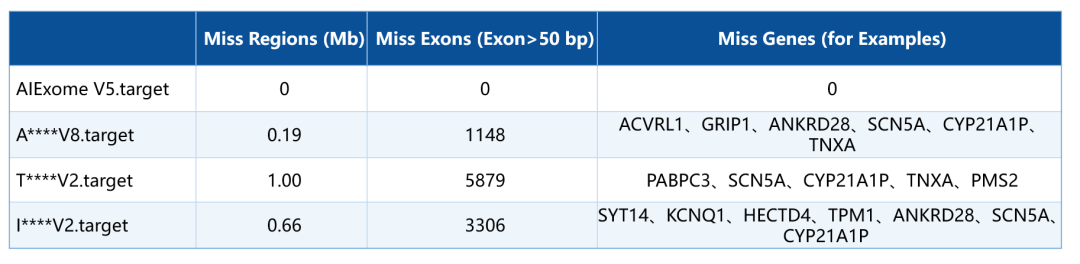
Scenario-based Solutions: Three Versions for Targeted Problem-Solving
Inherit Edition: Tackling Challenges in Genetic Disease Detection
The V5 Inherit Edition integrates over a decade of iGeneTech's experience in designing projects for monogenic genetic diseases, polygenic complex diseases, and rare diseases, providing cost-effective solutions for genetic disease gene detection and diagnosis.
01 Three-Tier Defense Network for CNV Detection:
First Layer of Assurance: Single-exon-level design for key genes such as Duchenne muscular dystrophy and thalassemia, improving detection rates;
Second Layer of Assurance: 10 kb high-density probe coverage for high-frequency pathogenic regions like 22q11.2 and 1p36;
Third Layer of Assurance: Genome-wide backbone probes with 200 kb intervals (50 kb encryption optional), enabling full coverage of CNVs from 1Mb to single-exon level.
This enhances the resolution of CNV detection for key genes to the single-exon level and supports genome-wide CNV detection of ≥ 1 Mb. Through special design for key genes, high-density probe coverage of large fragment deletion regions, and genome-wide backbone probe coverage, it helps improve the detection rate of various genetic diseases. The Genetic Whole Exome is pre-equipped with a genome-wide CNV backbone with 200 kb intervals (with 50/100 kb options available). Probes are evenly distributed, capable of covering intergenic regions and structurally complex regions.
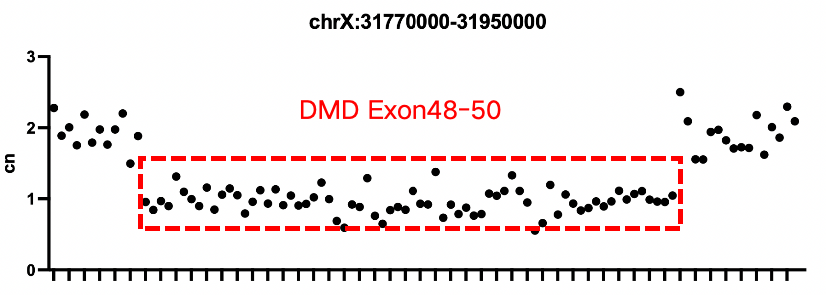
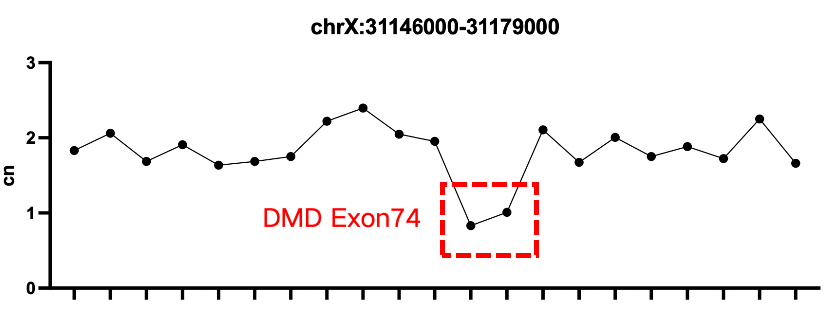
Verification of Positive Controls for Duchenne Muscular Dystrophy
Verification Protocol:
Sample 1: gDNA Reference Material 10 for Duchenne Muscular Dystrophy Gene Detection (heterozygous deletion of DMD exons 48–50)
Sample 2: gDNA Reference Material 14 for Duchenne Muscular Dystrophy Gene Detection (heterozygous deletion of DMD exon 74)
Library Preparation Kit: IGT® Enzyme Plus Library Prep Kit V3
Hybridization Capture Kit: TargetSeq One® Hyb & Wash Kit V3.0
Probe: V5 Inherit Edition
Conclusion:
According to the analysis results from CNV kit, a significant difference in sequencing depth can be observed in intron 47 and intron 50 of Sample 1, while Sample 2 shows a significant reduction in depth near exon 74.
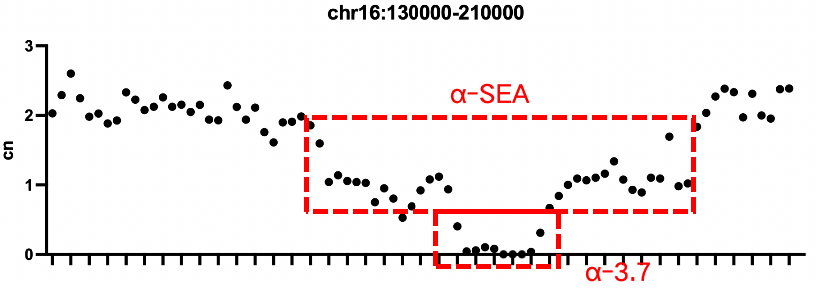
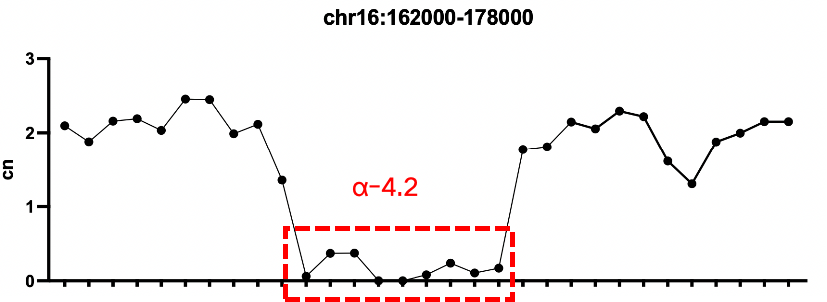
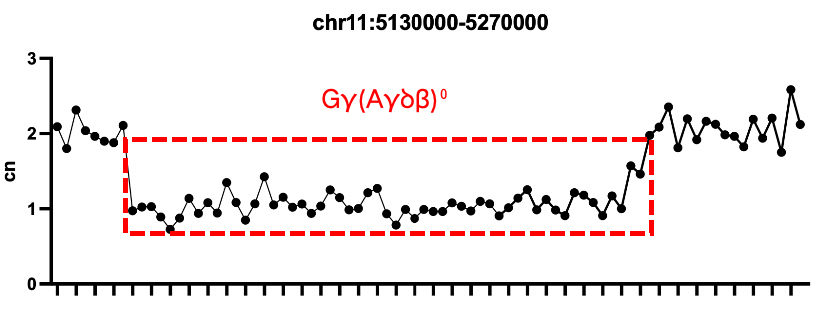
Verification of Positive Quality Control for Thalassemia
Verification Protocol:
Sample 1: Thalassemia α deletion gDNA Standard 3
Sample 2: Thalassemia α deletion gDNA Standard 6
Sample 3: Thalassemia β deletion gDNA Standard 2
Library Preparation Kit: IGT® Enzyme Plus Library Prep Kit V3
Hybrid Capture Kit: TargetSeq One® Hyb & Wash Kit V3.0
Probe: V5 Inherit Version
Conclusion:
Sample 1: -α3.7 homozygous deletion, -SEA heterozygous deletion
Sample 2: -α4.2 homozygous deletion
Sample 3: Gγ (Aγδβ)0 heterozygous deletion
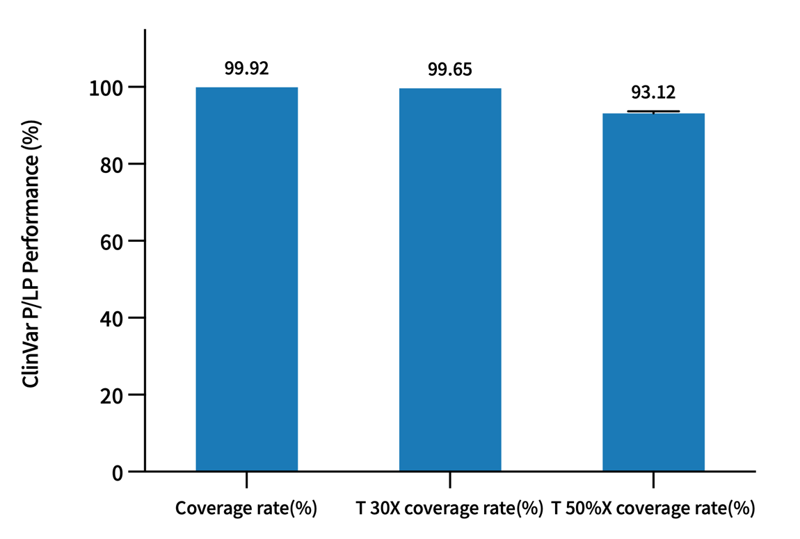
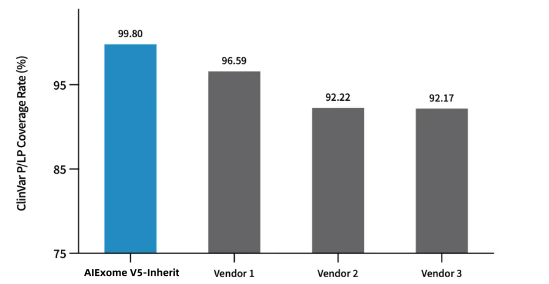
02 Gene Variation Harvester
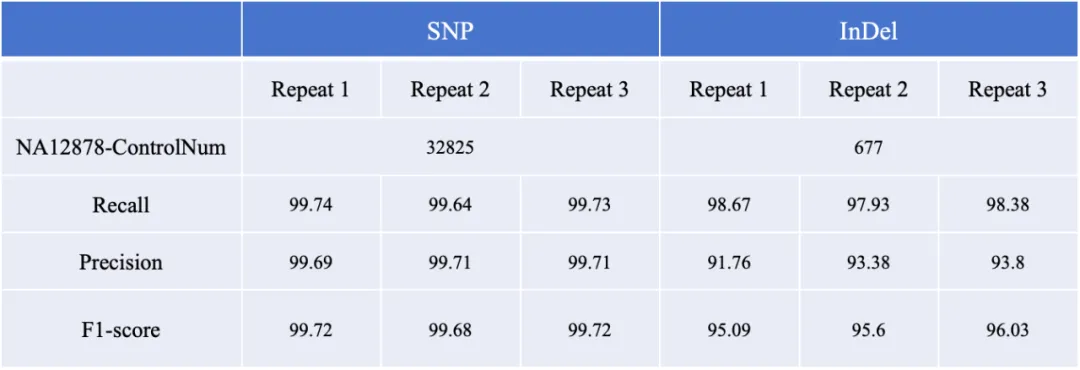
The coverage of P/LP (Pathogenic/Likely Pathogenic) loci in the ClinVar database reaches 99.8%. The detection performance of V5 Genetic Version probes was evaluated using the NA12878 standard cell line: the precision of SNP detection is as high as 99.74%, the recall rate reaches 99.71%, and the F1Score exceeds 99.72%; the F1Score for Indel detection is also higher than 96%.
03 Data indicators perform excellently with high mitochondrial depth
Spike-in full-length mitochondrial probes support high-depth sequencing of mitochondrial DNA, enabling the detection of mutations in all 37 mitochondrial genes, including point mutations, insertions/deletions, and heteroplasmy. Mitochondrial depth stabilizes at over 10,000×, making it suitable for the screening and diagnosis of mitochondrial-related diseases such as hereditary muscle weakness, hereditary optic neuropathy, mitochondrial encephalomyopathy, and deafness.
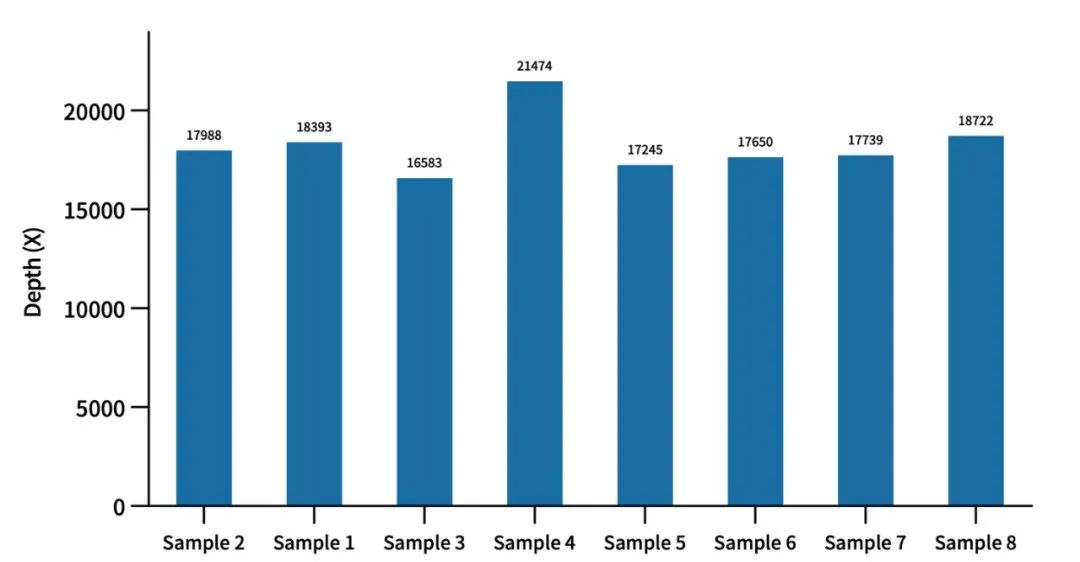
Evaluation of mitochondrial depth in actual measurement data of AIExome V5 Inherit Edition. Enzymatic digestion was performed for library construction using cell line quality control material, with a library input amount of 750ng and overnight hybridization. When the average depth of the target region of AIExome V5 Inherit Edition reaches 100×, the average mitochondrial depth can stably reach over 10000×.
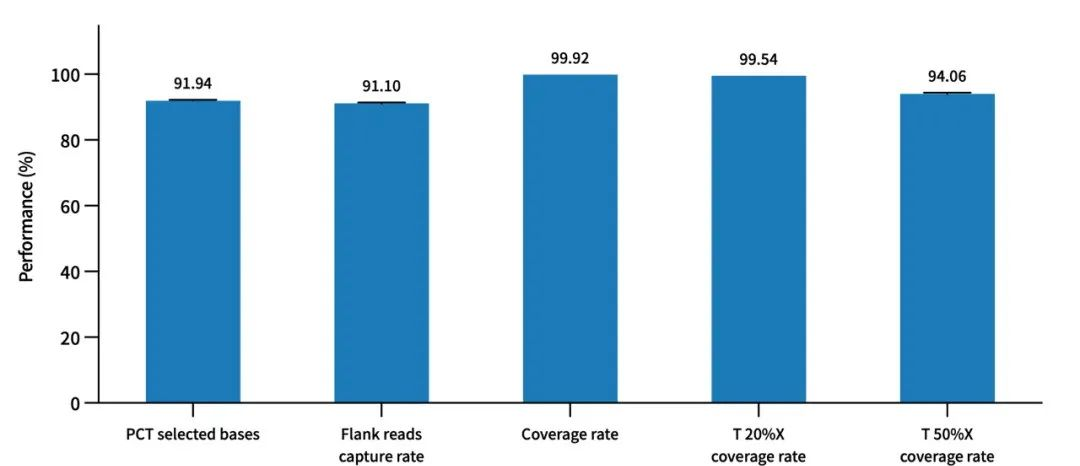
Performance of AIExome V5 Inherit Edition probes combined with TargetSeq one V3.0 in overnight hybridization capture. Enzymatic digestion was used for library construction with multi-person mixed standard (Promega, cat. G3041), with a library input amount of 750ng and overnight hybridization, followed by Illumina Novaseq6000 PE150 sequencing.
Tumor Edition: Precisely focusing on the field of precision oncology
01 All-in-one: Comprehensive detection capability
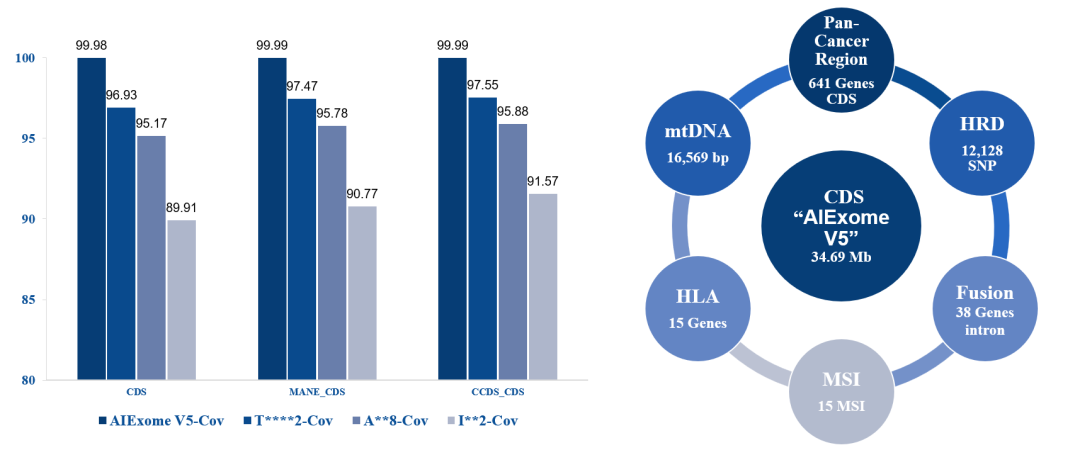
Figure 1. Coverage of CDS regions in various databases by AIExome® V5.
Figure 2. Coverage regions and biomarker coverage of AIExome® V5 Tumor Edition.
*RefSeq & refGene: The union of CDS from the latest RefSeq and refGene, with RefSeq version v20240129 and refGene version 20200817.
On the basis of covering over 99.95% of protein-coding regions, it newly includes 641 core oncogenes, 38 fusion hotspots, 15 MSI loci, and HLA immune repertoire. It enables detection of all variant types such as point mutations, CNV, Fusion, and MSI in a single tube, facilitating full-cycle management of targeted therapy, immunotherapy, and MRD monitoring.
02 Data Volume Optimization
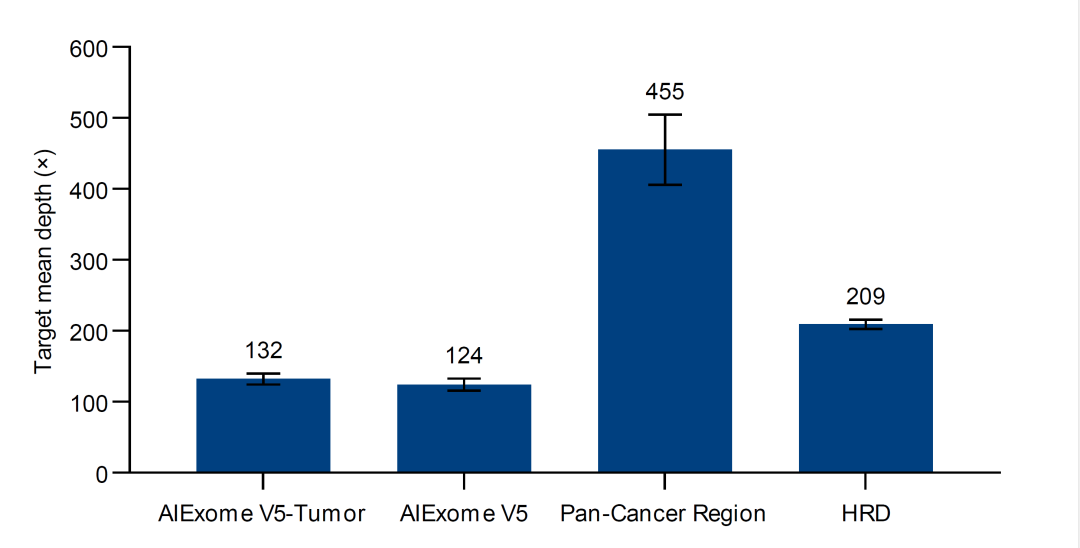
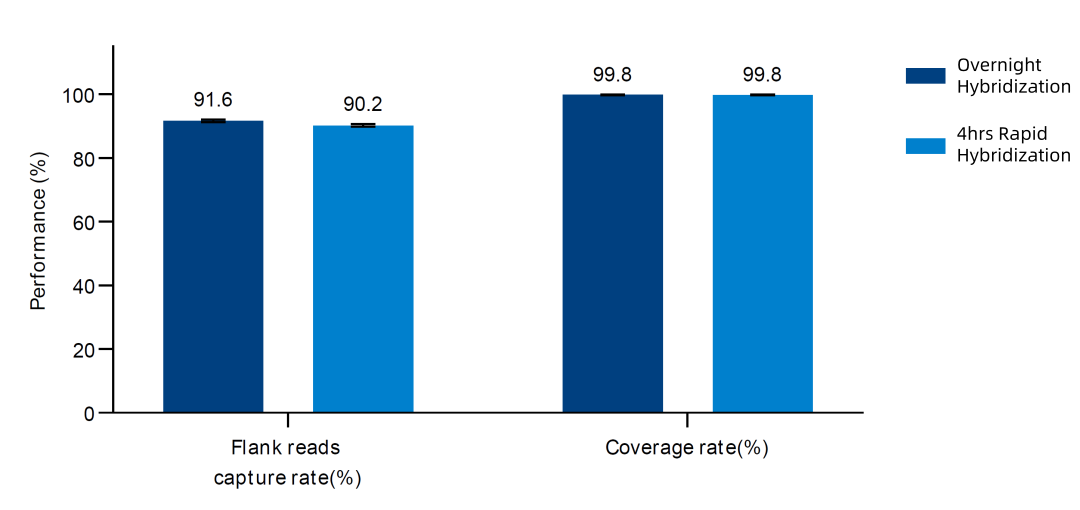
Figure 3. Statistics of depth ratios of different probe panels in AIExome® V5 Tumor Edition.
Figure 4. Capture performance of AIExome® V5 Tumor Edition probes combined with TargetSeq one V3.0 in 4hrs rapid hybridization and overnight hybridization.
The original depth difference regulation technology enables the depth of tumor-related genes to reach 4 times that of the whole exome region (with core gene depth of 400× under 10G data volume), saving 30G data volume compared with traditional whole exome.
03 "Golden" Partner for MRD Monitoring
Free customization of Tumor-informed MRD Panel, combined with UMI molecular tags to achieve ultra-high depth sequencing of 100,000×. The number of somatic mutation screenings has increased from 7 in traditional Panels to 126, comprehensively covering patient-specific somatic mutation sites, significantly improving detection accuracy and sensitivity.
Further Upgrade of Customized Services
680K Independent Probe Synthesis Platform: Supports regional encryption/customization within 24 hours, creating exclusive whole exome products for research teams;
High-Quality Product Services: With 11 years of experience in probe design, synthesis, and development of supporting reagent products, we can customize personalized whole exome sequencing products and solutions according to different customer needs.

 CN
CN