The CRISPR-Cas system is an important tool in gene editing research, and the quality of sgRNA libraries (throughput, uniformity, accuracy) is crucial to the success of screening experiments (such as whole-genome screening, drug target discovery, and agricultural breeding).
Traditional sgRNA synthesis methods face the following challenges:
l High-throughput demand: Whole-genome screening requires the synthesis of a large number of sgRNA sequences, and traditional methods are inefficient.
l Uniformity issues: Uneven concentrations of sequences in the library lead to the omission of some targets (false negatives), affecting the reliability of screening results and causing resource waste.
l Off-target risk: Errors in sgRNA sequence synthesis are one of the main sources of off-target effects in experiments.
To address these, iGeneTech has launched the Ignite 3.0 high-throughput DNA synthesis platform, dedicated to providing high-quality sgRNA library synthesis solutions.
I. Four Advantages of iGeneTech's Ignite 3.0 High-Throughput DNA Synthesis Platform
1. High-throughput synthesis:
l Supports the synthesis of Oligo Pools at scales ranging from 4k to 650k (with varying complexity).
l Stably supports synthesis lengths of 200 nt.
l A single synthesis run can cover full-scale needs (for example, approximately 20,000 protein-coding genes in humans, with 3-5 sgRNAs designed for each gene).
This reduces the inefficient mode of "batch synthesis and repeated waiting" and significantly shortens the library construction cycle.
2. High sequence uniformity:
l Library coverage exceeds 99.9%.
l The minimum yield guarantee for a single sequence is ≥ 0.5 fmol.
l The 95%/5% abundance quantile ratio is as low as 1.82.
This ensures that each sgRNA can be equally expressed and detected in downstream experiments, reducing target omission caused by abundance differences.

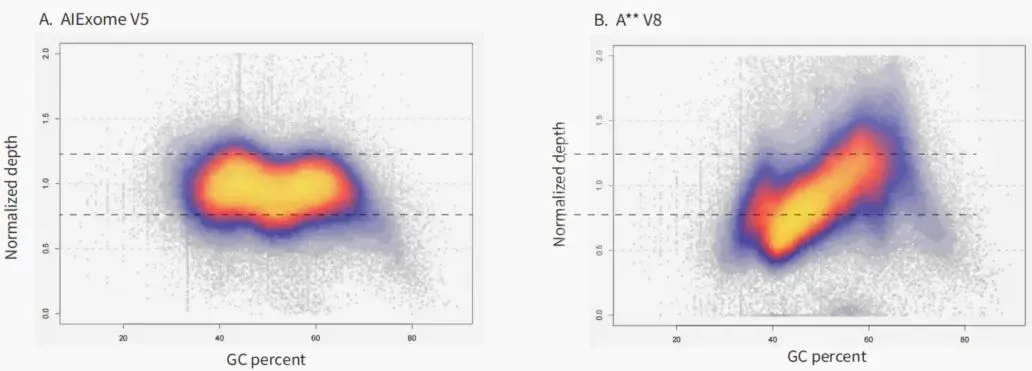
Figure 1. Oligo Pool Uniformity Performance of the Ignite 3.0 Platform (Illustration: Oligo Pool Sequencing Depth Distribution Map of the Ignite 3.0 Platform, showing high uniformity of 30,000 200 nt sequences with a 95%/5% ratio of 1.82)
3. Low synthesis error rate:
l The average single-base error rate (substitutions, insertions, and deletions) is controlled at approximately 0.1%.
l UMI molecular tag verification shows that the comprehensive error rate of 3,000 130 nt sequences is only 0.1629%.
This ensures high accuracy of sgRNA sequences, thereby helping to reduce the risk of off-target effects in experiments from the source.
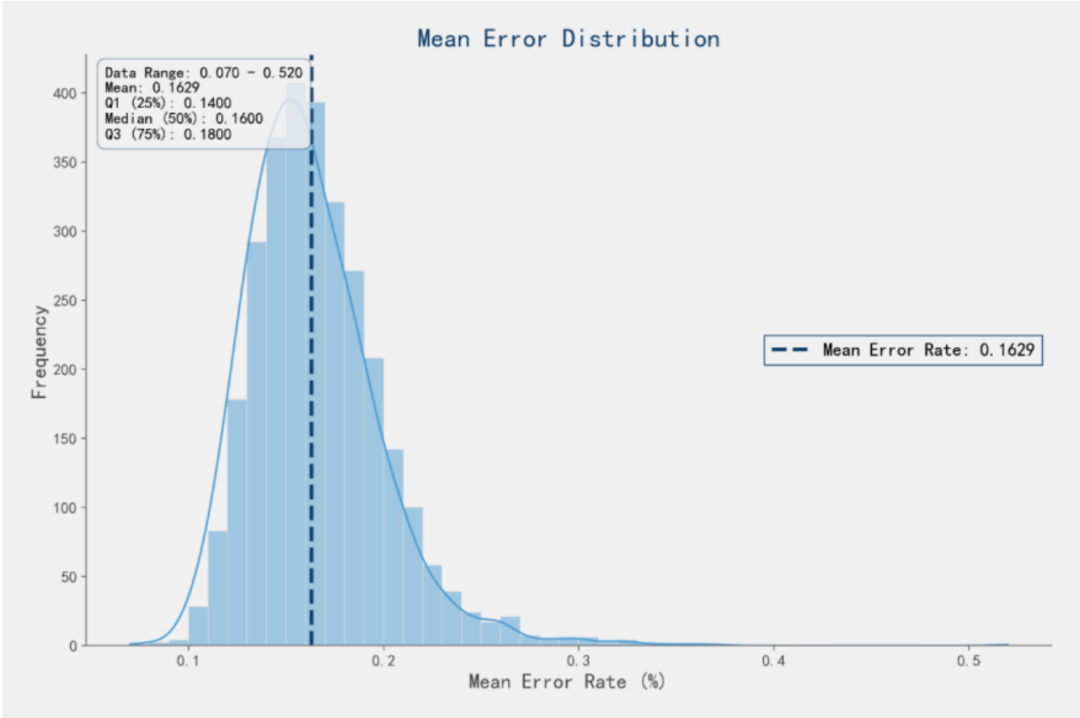
Figure 2. Oligo Pool Synthesis Error Rate Performance of the Ignite 3.0 Platform (Illustration: Oligo Pool Synthesis Error Rate Distribution Map of the Ignite 3.0 Platform, with an average error rate of 0.1629%)
4. Fast delivery:
l The standard delivery time for the entire process of customized Oligo Pool synthesis is 5 working days (from sequence submission to delivery of DNA dry powder).
l Each synthesis chip includes a dedicated quality control (QC) Pool to monitor synthesis quality in real time.
II. Dual-Technology Detection System for Off-Target Effects
Off-target effects are a core risk in basic research and clinical translation of CRISPR technology. Off-target detection needs to meet both high-throughput and high-sensitivity requirements.
To address this, iGeneTech offers two complementary solutions:
1. TargetSeq® Hybrid Capture Sequencing:
l Uses customized probes to simultaneously enrich target sites and potential off-target sites (hundreds of them).
l Combined with UMI tags, the detection sensitivity is better than 0.1%.
l Applicability: Precise detection of target sites and high-risk off-target regions, suitable for preclinical research.
2. MultipSeq® Multiplex Amplicon Sequencing:
l Uses high-throughput designed primers to simultaneously detect target and off-target sites (hundreds of them).
l The experimental process is simplified, with a sensitivity better than 1%.
l Applicability: Suitable for large-scale off-target site screening in basic research.
III. Multi-Field Application Expansion
The Oligo Pools synthesis capability of iGeneTech’s Ignite 3.0 platform can meet the needs of multiple fields such as basic scientific research, pharmaceutical R&D, and agricultural biotechnology, and provides customized solutions:
l Mutant library construction: Customizes Pools containing specific variations such as point mutations, insertions/deletions, for gene function research, protein engineering optimization, and crop trait improvement screening.
l High-throughput reporter gene analysis (MPRA): Customizes Pools covering regulatory elements such as promoters, enhancers, and disease-related SNPs, enabling batch analysis of element activity for studying gene regulatory mechanisms or drug target validation.
l Fluorescence in situ hybridization (FISH) probe preparation: Rapidly customizes probe libraries targeting specific genes or chromosomal regions, eliminating the need for long-fragment cloning.
iGeneTech focuses on providing customers with high-quality Oligo Pools synthesis and high-sensitivity targeted capture sequencing technology services to meet the key needs of cutting-edge research fields such as gene editing.

 CN
CN