Detection of fusion genes at the RNA level
Fusion genes often serve as molecular markers for tumorigenesis, and are of great significance for clinical diagnosis, drug therapy, and prognosis. They may be closely related to the occurrence and development of various cancers, and may also be potential drug targets. In the detection of tumor fusion genes, fusion breakpoints usually occur in long intronic regions.
There are several intractable problems in detecting fusion genes at the DNA level:
1. Probes need to cover lengthy intronic regions that contain a large number of repetitive sequences;
2. High GC content is unfavorable for probes to capture fragments of the target region;
3. The introns of different genes have similar repetitive sequences, a feature that hinders accurate sequence alignment and affects detection accuracy;
4. Complex transcriptional or post-transcriptional splicing processes may interfere with gene fusion.
Compared with the DNA level, fusion genes at the RNA level are manifested as junctions between exons of two adjacent genes, with relatively fixed fusion sites. This provides an excellent advantage for the precise design of probes.
Therefore, based on the sequence characteristics of fusion genes, detecting fusion genes at the RNA level is easier to achieve than at the DNA level.
iGeneTech has currently developed RNA-based capture products, including those for solid tumor fusions, hematological tumor fusions, and RNA exome sequencing, meeting diverse needs in different scenarios.
01 Differences between RNA capture and DNA capture
l Samples: RNA capture requires extracting total RNA from tissues, FFPE samples, and blood samples; for DNA samples, simply extracting DNA is sufficient.
l Library construction: For the extracted total RNA, RNA library construction kits are needed, and since the capture process will be performed subsequently, the procedure for rRNA regions is unnecessary; DNA samples use DNA library construction kits.
l Capture: The capture process is the same (in special scenarios such as pathogenic microorganism capture, it is recommended to add rRNA blocking reagents).
l Application scenarios: The RNA level is mainly used for detecting gene fusions and gene expression, etc.; the DNA level is used for detecting variations such as SNV, InDel, CNV, Fusion, and SV.
02 iGeneTech's RNA capture design scheme
A. In probe design for detecting fusion genes at the RNA level, a 3× tiling design is first carried out based on all transcript sequences of the target genes. Meanwhile, while ensuring that each transcript achieves 3× tiling coverage, redundant probes that repeatedly cover the same segments are removed.
B. Sufficient coverage of exon junction regions is fully guaranteed; during design, additional probes are supplemented in exon splicing regions to improve the detection rate of fusion genes.
C. For known fusions, especially low-frequency fusion variants, mutant probes can be added according to the fusion sequences to enhance the detection rate.

Figure 1. Probe design method for detecting fusion genes at the RNA level
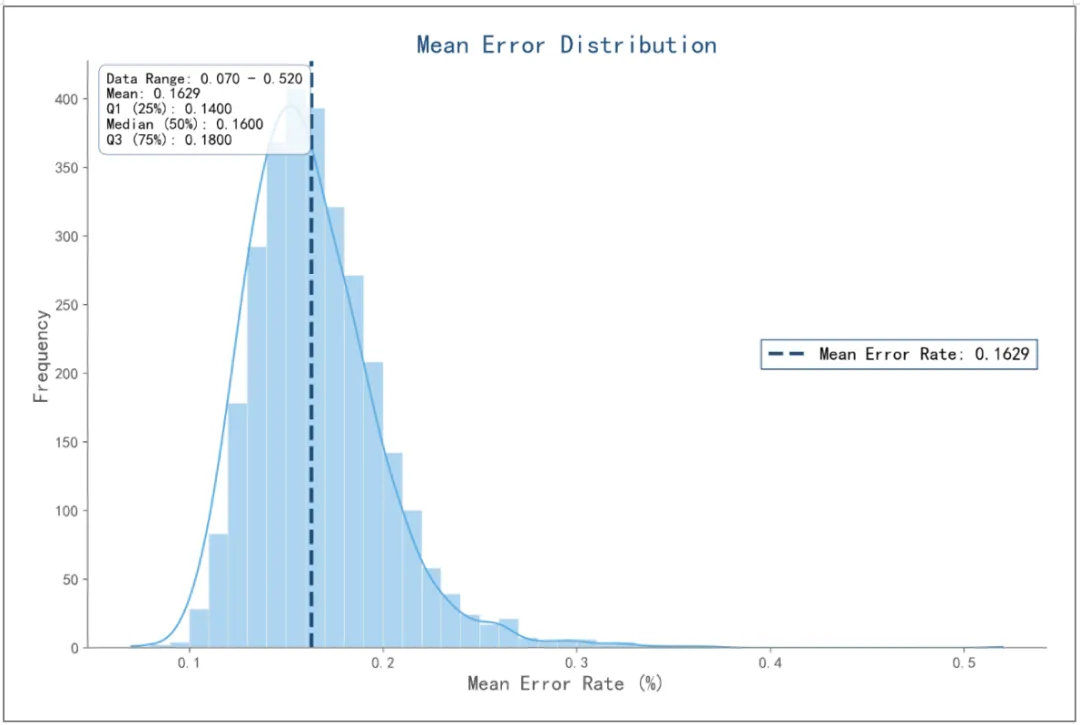
03 RNA Capture Performance Evaluation Protocol
l Capture and sequencing are conducted on gDNA samples to assess the coverage and uniformity performance of the RNA capture panel.
l Capture and sequencing are performed on RNA samples to evaluate the capture efficiency of the capture library. At this point, the characteristics of differential gene expression in RNA samples also exert a significant impact on the data regarding coverage and uniformity.
l Detection of fusion events is carried out on known fusion RNA samples
04 iGeneTech RNA Capture Pre-defined Products


 CN
CN