Recent national-level surveillance data indicates that influenza activity has been on a continuous rise in both northern and southern China, and a multi-region upward trend has also been observed globally. In the 45th week of 2025, over 21,000 specimens from influenza-like cases were tested nationwide, with a positivity rate of 24.7%. The A (H3N2) subtype is the absolutely dominant circulating strain, accounting for more than 98% of all positive cases detected in both northern and southern China.
Meanwhile, a total of 621 outbreaks of influenza-like cases have been reported across the country, showing a significant increase compared with the same period in previous years. Outbreaks have occurred frequently in crowded places such as schools and workplaces. Globally, the influenza positivity rate has exceeded 30% in many countries, especially those in Asia, Africa and parts of Europe. The A (H1N1)pdm09 and A (H3N2) subtypes are co-circulating in various regions. At the same time, highly pathogenic avian influenza cases have been continuously reported in multiple countries, and the risk of cross-species transmission cannot be ignored.
Overall, influenza viruses are showing a trend of high prevalence, high positivity rate and multi-point outbreaks both domestically and internationally, which imposes higher requirements on the capabilities of rapid influenza virus detection, drug resistance monitoring and whole-genome tracing.
Based on TargetSeq® liquid-phase probe capture technology, iGeneTech has developed the Influenza A/B/C Virus Panel, a whole-genome liquid-phase capture kit for influenza viruses. Taking the full-length genomic sequences of influenza A (taxid: 11320), B (taxid: 11520), and C (taxid: 11552) viruses from NCBI over the past 20 years as target regions, the kit is designed with a total of 33,609 probes. It achieves high-density coverage of highly variable viral regions, enables accurate typing of influenza viruses, and ensures more precise detection of emerging mutations.
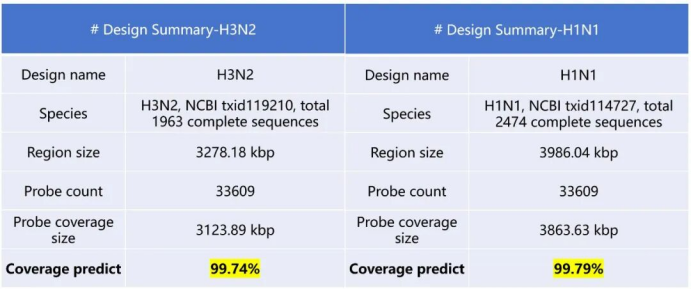
For currently circulating influenza virus strains such as H3N2 and H1N1, the Influenza A/B/C Virus Panel achieves over 99.7% coverage of sequences from relevant subtypes and lineages in the past five years. Additionally, the probe hybridisation capture technology enables better capture and sequencing of emerging mutations generated during the rapid spread of viruses. It provides higher-resolution monitoring and tracing capabilities for public health surveillance systems, assisting in evaluating vaccine match and future epidemic trends.
Whole-genome capture sequencing of influenza viruses upgrades influenza surveillance from "whether influenza exists" to "which strain it is, where it comes from, and whether it will mutate." It is a core tool for enhancing regional CDC capabilities, strengthening outbreak response, and optimizing prevention strategies.
Product Info
Product Name | Speci. | Cat. No |
Influenza A/B/C Virus Panel | 16 rxn / 96 rxn | PH2000051/ PH2000052 |
IGT® RNA Pathogen Microbial Library Prep & Capture Kit(Illumina) | 16 rxn | C11371 |
IGT® RNA Pathogen Microbial Library Prep & Capture Kit(MGI) | 16 rxn | C11441 |
Magnetic Beads Based Pathogen DNA / RNA Co-Extraction Kit | 50 rxn | E10021 |
Magnetic Bead Method Pathogenic DNA/RNA Extraction Kit | 50 rxn | E20011 |

 CN
CN


