In August 2022, IGT™ ssDNA Library Prep Kit, the brand-new single-stranded DNA library preparation kit by iGeneTech, was launched. The kit can be used for methylation library and genomic library construction from 1 ng to 50 ng of starting DNA. The constructed library can be further used for target region hybrid capture sequencing or direct sequencing. During the construction of methylation libraries, bisulfite treatment can cause significant DNA damage, fragmentation, and denaturation. Moreover, low-quality or severely degraded DNA contains a large amount of single-stranded DNA. The IGT™ ssDNA Library Prep Kit allows adaptor ligation to single-stranded DNA, significantly improving the utilization of the original DNA molecules and library complexity. This provides a significant advantage in constructing methylation and genomic libraries from low-input samples.
Background
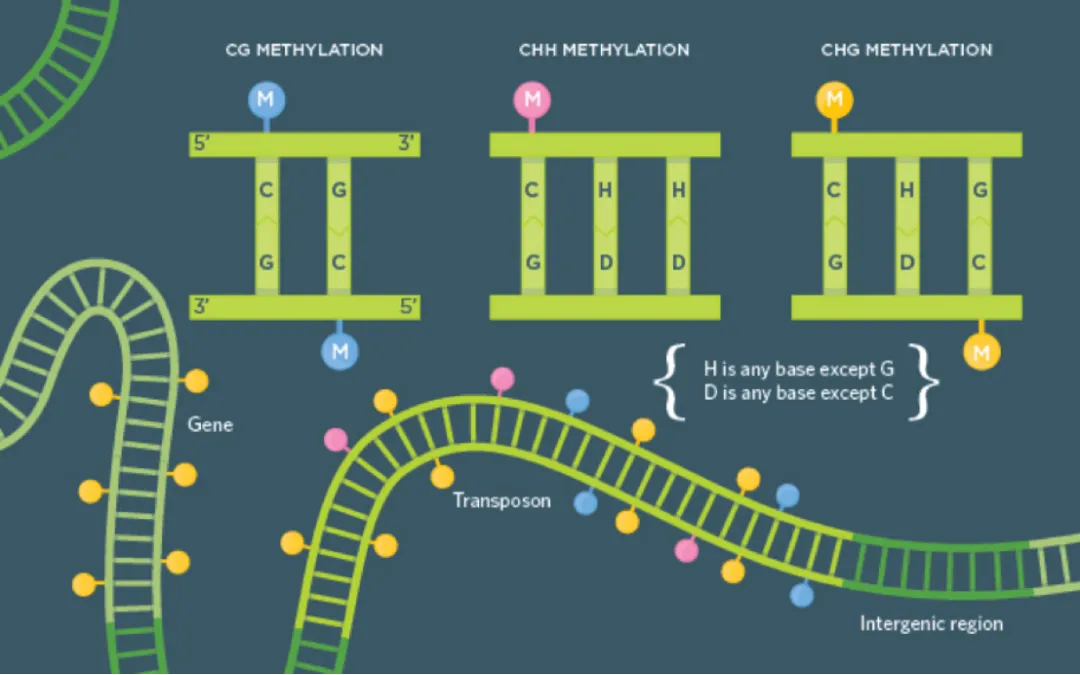
DNA methylation generally refers to the 5-methylcytosine (5mC) modification on DNA, which is the most abundant epigenetic modification on DNA and one of the most extensively studied. There is a significant difference in DNA methylation states between normal cells and tumor cells. For example, in normal cells, CpG islands in the promoter regions of many tumor suppressor genes are in a low methylation state, while in tumor cells, these CpG islands are heavily methylated, leading to decreased expression of tumor suppressor genes. Changes in methylation states can often be detected in the early stages of tumorigenesis, giving DNA methylation the potential as an early screening biomarker for tumors. Additionally, DNA methylation exhibits significant tissue specificity, enabling tissue traceability by detecting DNA methylation states.
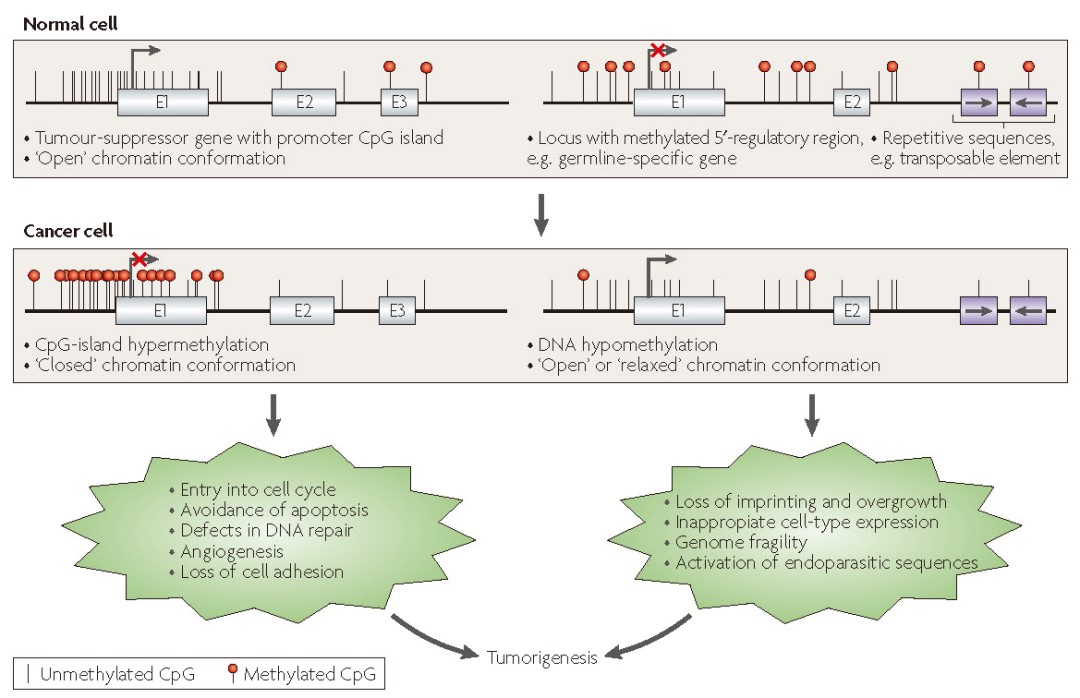
Figure 1. DNA methylation levels in tumor and normal cells[1] However, the abundance of early-stage cancer ctDNA in vivo is low. Current technical challenges in free nucleic acid DNA methylation detection include cytosine conversion rate and the utilization rate of the original DNA molecules. Bisulfite treatment can achieve a stable and efficient cytosine conversion rate. However, in traditional Pre-BS library preparation, where adaptors are ligated to double-stranded DNA before bisulfite treatment, significant DNA damage and fragmentation occur due to bisulfite treatment. Most DNA molecules with ligated adaptors cannot be amplified due to fragmentation, leading to low utilization rates of the original DNA molecules, severely limiting their application in cfDNA. The Post-BS library preparation strategy can achieve both high cytosine conversion rates and high utilization rates of the original DNA molecules. Based on the Post-BS library preparation strategy, iGeneTech launches the IGT™ ssDNA Library Prep Kit, a universal single-stranded DNA library preparation kit. In this method, DNA is first treated with bisulfite and then ligated with adaptors, allowing fragmented DNA molecules due to bisulfite treatment to be utilized, significantly increasing library complexity and data validity, making it especially suitable for low-input samples like cfDNA.
IGT™ ssDNA Library Prep Kit Methylation Library Construction Workflow
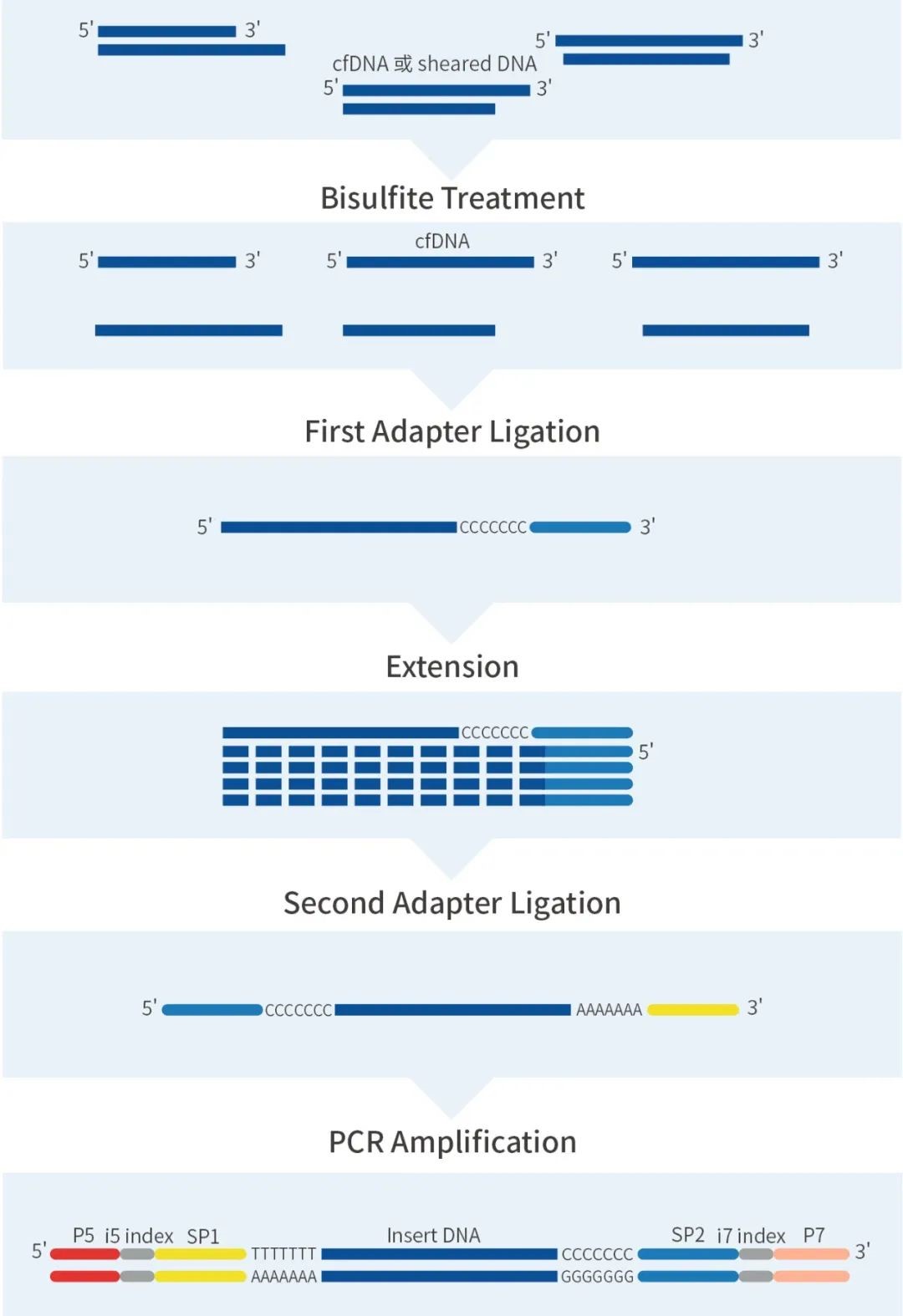
Figure 2. IGT™ ssDNA Library Prep Kit Methylation Library Construction Workflow
Product Benefits
Suitable for low-input samples like cfDNA and FFPE DNA; library input can be as low as 1 ng;
Utilizing the Post-BS library preparation strategy significantly increases the utilization of DNA templates during methylation library construction;
Effectively increases library complexity and sequencing data validity;
Compatible with whole-genome methylation sequencing, target region methylation sequencing, whole-genome sequencing, and target region hybrid capture sequencing.
Methylation Library Sequencing Data Display
Performance of Different Input Samples
The IGT™ ssDNA Library Prep Kit, in combination with the BisCap® hybrid system, is suitable for different input samples, showing excellent data performance.
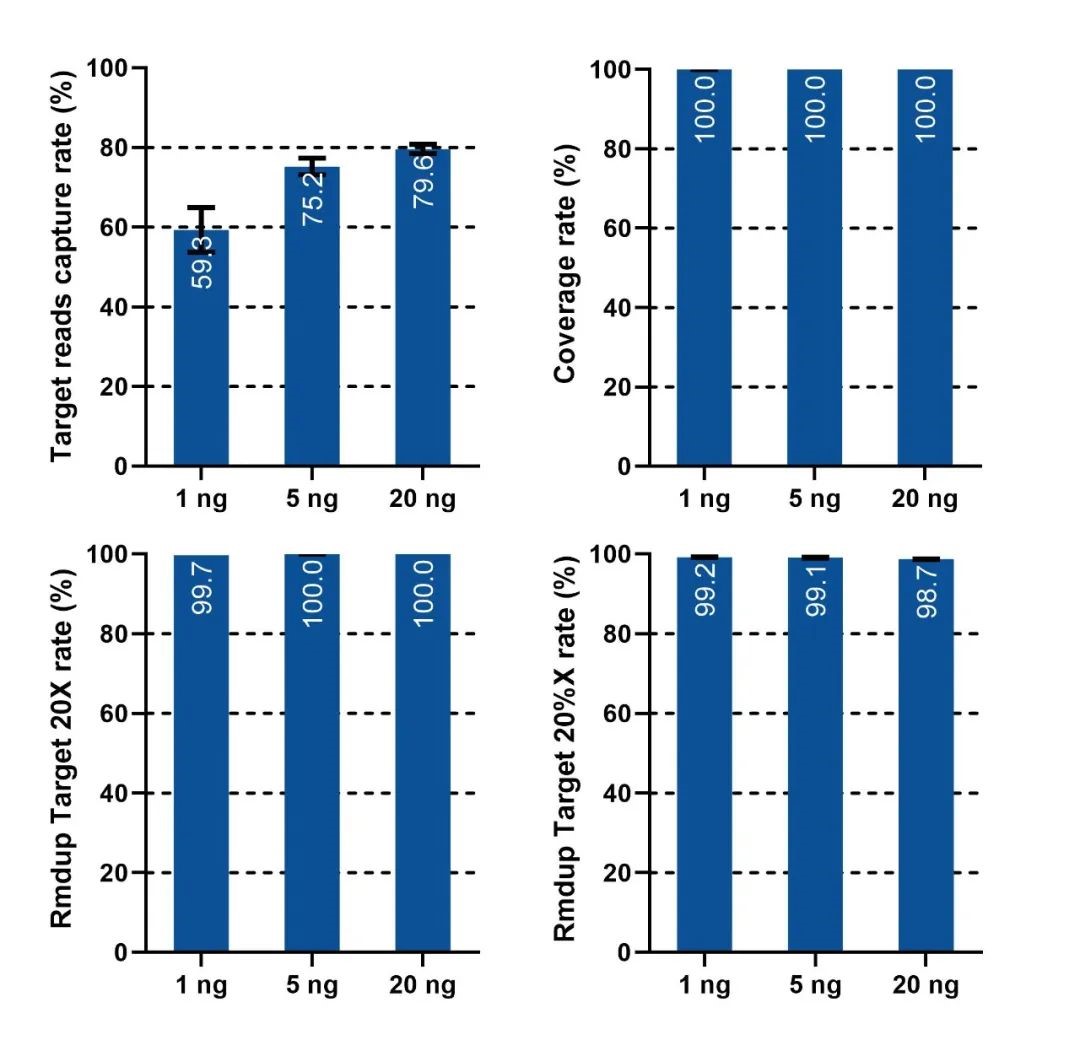
Figure 3. Capture data performance of the IGT™ ssDNA Library Prep Kit with different input amounts. Single-stranded libraries were prepared with 1 ng, 5 ng, and 20 ng of cfDNA (sample is NA12878 cell line gDNA, sonicated to simulate cfDNA). Library construction was performed using the IGT™ ssDNA Library Prep Kit and IGT™ ssDNA Adapter & UDI Primers (for Illumina), and captured using a 123 kb Panel, sequenced on the NovaSeq 6000 platform with PE150, generating an average of 1.8 Gb of sequencing data.
Very High Cytosine Conversion Rate
The IGT™ ssDNA Library Prep Kit uses bisulfite transformation during library preparation, achieving a cytosine conversion rate of over 99.2%.

Figure 4. The conversion rate performance of the IGT™ ssDNA Library Prep Kit with different input amounts. Bisulfite transformation libraries were prepared with 1 ng, 5 ng, and 20 ng of cfDNA (sample is NA12878 cell line gDNA, sonicated to simulate cfDNA), and library construction was performed using the IGT™ ssDNA Library Prep Kit and IGT™ ssDNA Adapter & UDI Primer (for Illumina), captured using a 123 kb Panel, and sequenced on the NovaSeq 6000 platform with PE150, generating an average of 1.8 Gb of sequencing data.
High Utilization Rate of Original DNA Molecules
Under the same amount of sequencing data, single-stranded library input is only 1/10 of double-stranded library input, with single-stranded library having a higher effective depth post-deduplication.

Figure 5. Effective depth performance of the IGT™ ssDNA Library Prep Kit and double-stranded DNA library preparation kit. Samples were NA12878 cell line gDNA, with library construction performed with the input of 5 ng, 20 ng, 50 ng, and 200 ng. 5 ng and 20 ng library constructions used the IGT™ ssDNA Library Prep Kit and IGT™ ssDNA Adapter & UDI Primer (for Illumina), and 50 ng and 200 ng library constructions used the IGT™ Methyl Fast Library Prep Kit v2.0 and IGT™ Methyl Adapter & UDI Primer (for Illumina), captured using a 123 kb Panel, and 1 Gb of data was extracted for effective depth analysis.
Excellent Reproducibility of Methylation Levels Post-Library Capture
The IGT™ ssDNA Library Prep Kit, combined with the BisCap® hybrid system, shows excellent reproducibility of methylation levels between replicate samples.
A:
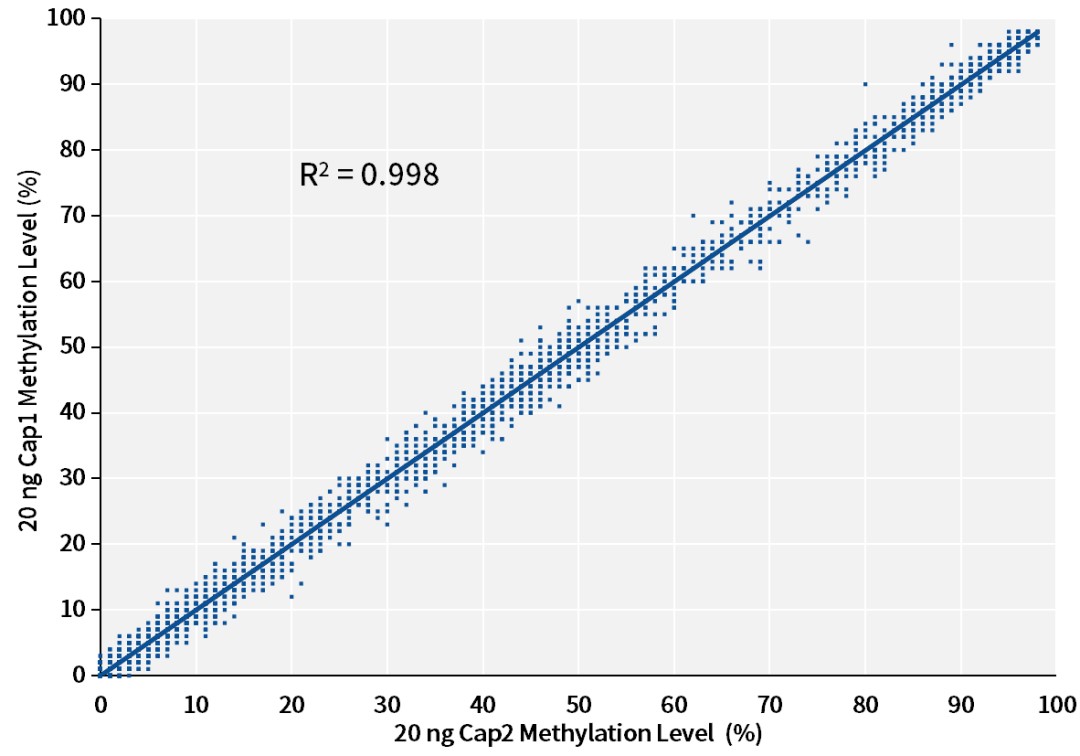
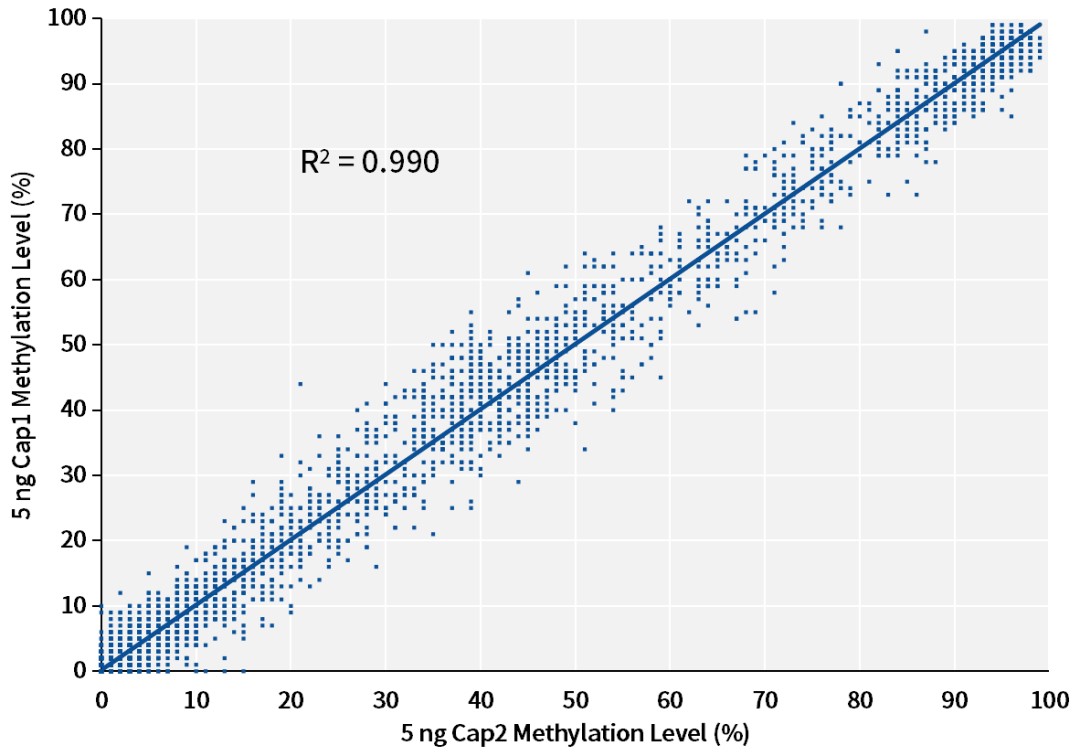
B:
C:
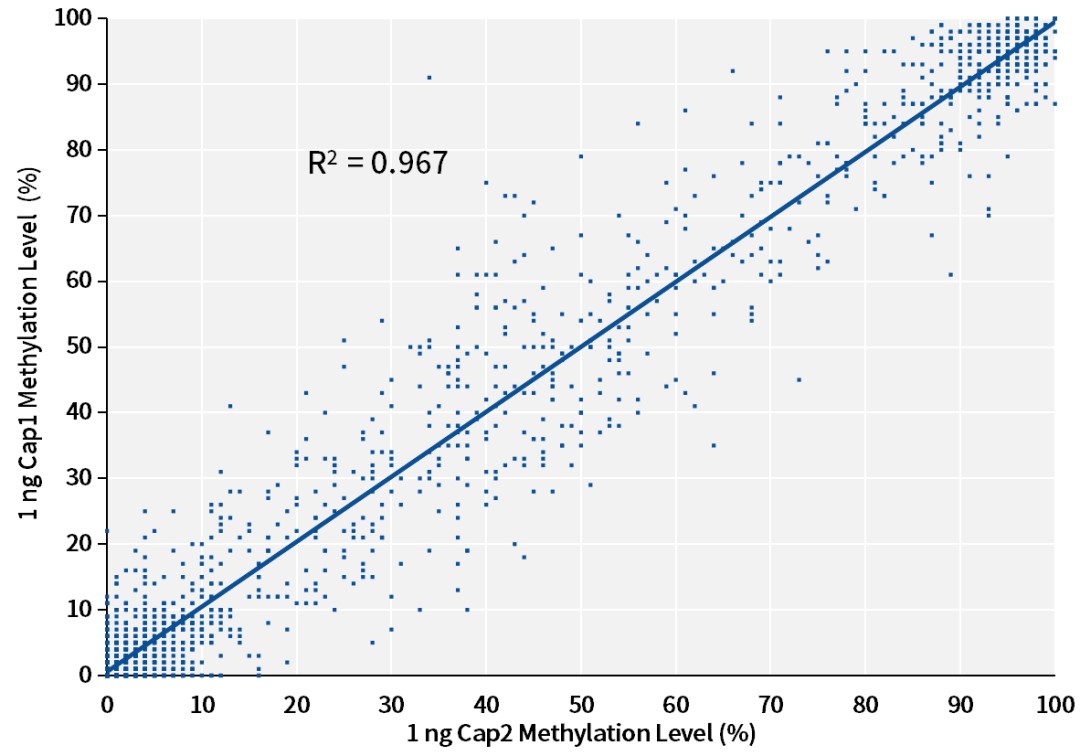
Figure 6. Reproducibility of methylation levels with different DNA input amounts using the single-stranded DNA library preparation kit. Single-stranded libraries were prepared with the input of A: 20 ng, B: 5 ng, C: 1 ng cfDNA (sample is NA12878 cell line gDNA, sonicated to simulate cfDNA), using the IGT™ ssDNA Library Prep Kit and IGT™ ssDNA Adapter & UDI Primer (for Illumina), captured using a 123 kb Panel, sequenced on the NovaSeq 6000 platform with PE150, and analyzed for reproducibility of methylation levels at CpG sites in different batches of the same sample capture experiments.
Excellent Consistency of Methylation Levels Between Single-Stranded and Double-Stranded Libraries
The methylation levels detected in single-stranded libraries are highly consistent with those in double-stranded libraries, with an R2 of 0.99.
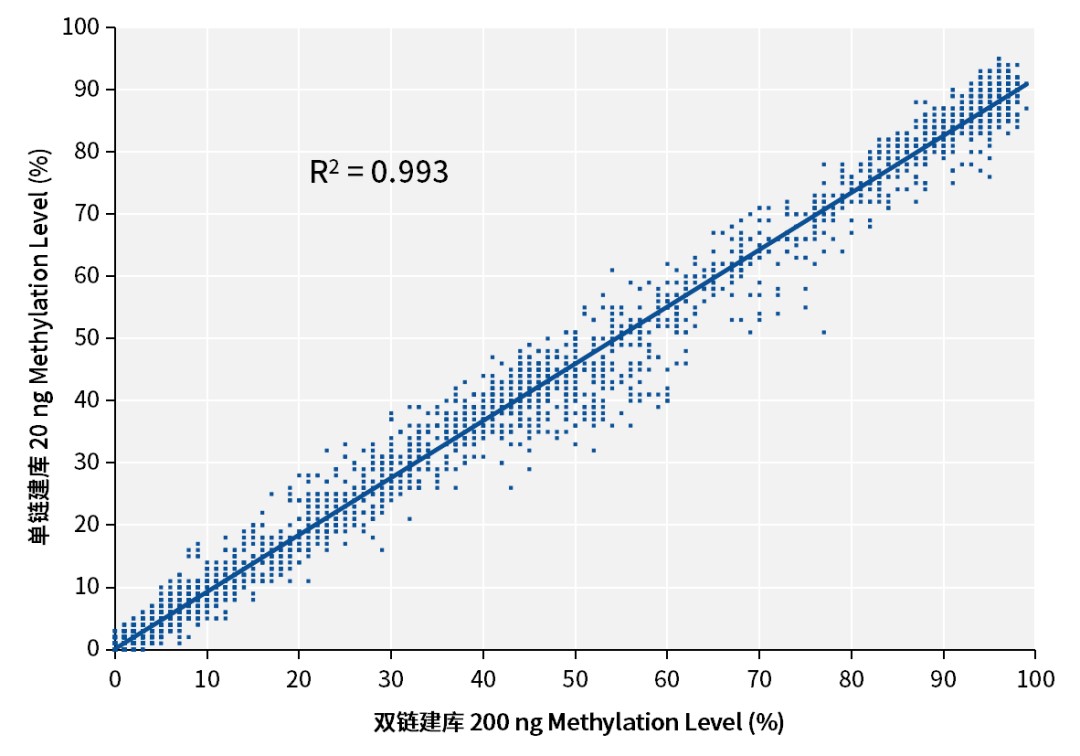
Figure 7. Correlation of methylation levels between double-stranded and single-stranded library preparation technologies. Libraries were prepared with the input of 20 ng cfDNA (sample is NA12878 cell line gDNA, sonicated to simulate cfDNA) and 200 ng (sample is NA12878 cell line gDNA) by sonication. The 20 ng input sample libraries were prepared with the IGT™ ssDNA Library Prep Kit and IGT™ ssDNA Adapter & UDI Primer (for Illumina), and the 200 ng input sample libraries were prepared with the IGT™ Methyl Fast Library Prep Kit v2.0 and IGT™ Methyl Adapter & UDI Primer (for Illumina), captured using a 123 kb Panel, and sequenced on the NovaSeq 6000 platform with PE150. Methylation level correlation between single-stranded and double-stranded library preparation technologies was analyzed at the same sequencing depth (1000×).
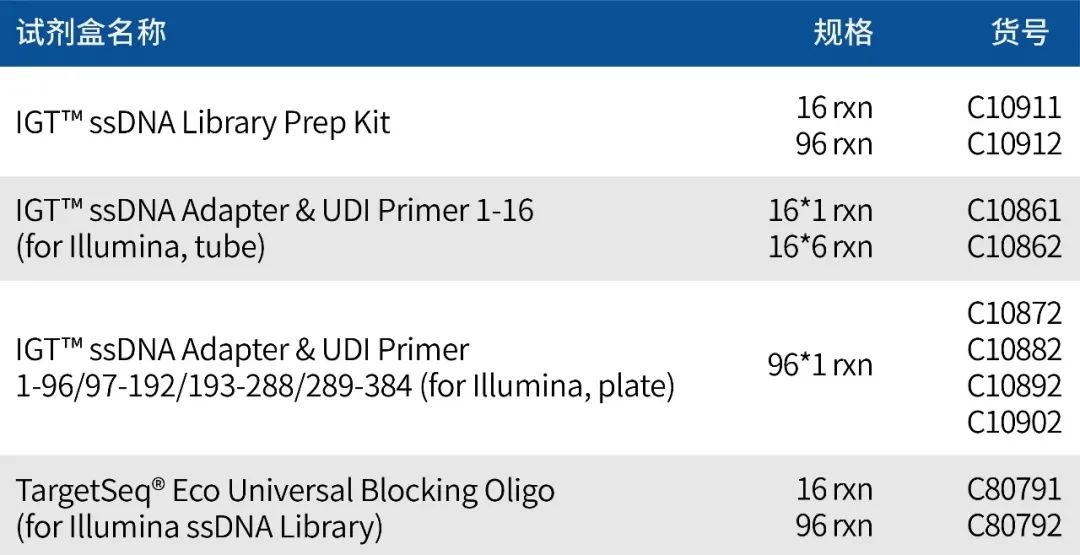
Product Information
About iGeneTech
iGeneTech is a high-tech Chinese enterprise focused on developing and supplying target gene "reading" and "writing" technology solutions. It possesses three fundamental technology platforms with independent core intellectual property rights: NGS probe hybridization, multiplex PCR, and high-throughput DNA synthesis. iGeneTech has been certified by multiple compliance production quality management systems, including ISO 13485 and ISO 9001, with a 2500㎡ GMP production workshop and testing laboratory. It provides customized development of gene capture products, NGS IVD product CDMO services, targeted sequencing laboratory solution supply, rapid NGS sequencing services, and large-scale DNA synthesis services for fields such as medical health, agriculture, microbiology, and academic research. iGeneTech is dedicated to being an excellent partner for genetic testing companies, hospital precision medicine centers, research institutions, synthetic biology companies, and drug development institutions, creating a new leading engine in the gene industry. iGeneTech has provided hundreds of prefabricated gene capture products and high-performance reagent components, over 1500 high-standard personalized gene capture products for nearly a thousand customers in China, and core raw materials for dozens of NGS IVD products in progress or under review.

 CN
CN