In the field of tumor genetic testing, whole exome sequencing (WES) enables "panoramic" exploration, facilitating in-depth insight into tumor genetic profiles, accurately determining tumor types and molecular subtypes, and helping develop personalized treatment plans for patients.
Leveraging the advantages of AIExome® V5 Core Edition—including comprehensive coverage, high-efficiency capture, and breakthrough uniformity metrics—iGeneTech has launched a new product tailored for tumor genetic testing scenarios: AIExome V5-Tumor Edition. With its multi-dimensional performance advantages, this product precisely focuses on the precision oncology field, better meeting the needs of clinical testing and scientific research.
Highlight 1: All-in-one Comprehensive Detection
All-around coverage for tumor testing scenarios—AIExome V5-Tumor, based on covering over 99.95% of protein-coding regions from databases such as CDS (RefSeq & refGene)*, MANE, and CCDS, additionally includes detection regions for Fusion, MSI, HLA, HRD, and mtDNA. It achieves multi-variant detection needs in a single probe tube and one-time testing, truly realizing All-in-one.
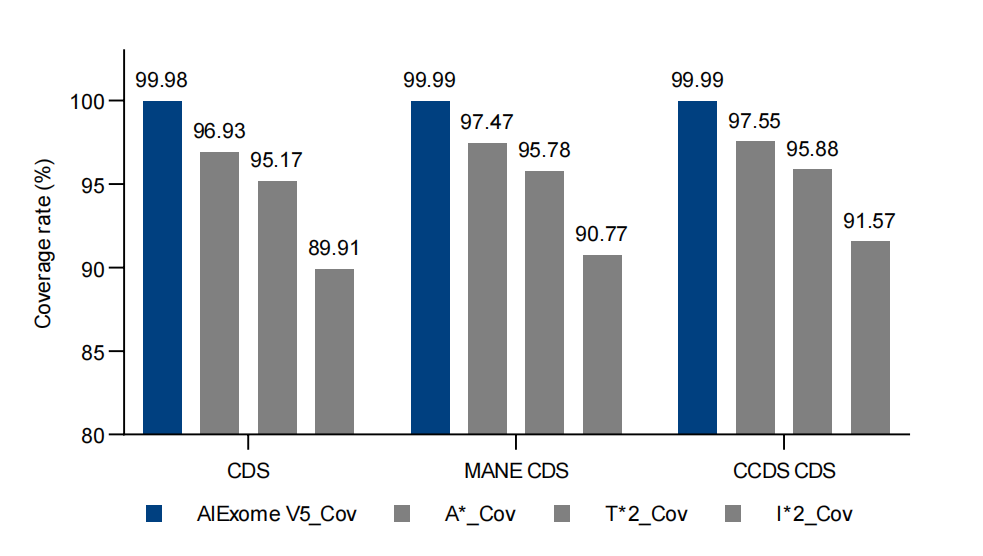
Figure 1. Coverage of CDS regions in various databases for AIExome V5 compared with competing products. *RefSeq & refGene: Union of CDS from the latest RefSeq (version v20240129) and refGene (version 20200817).
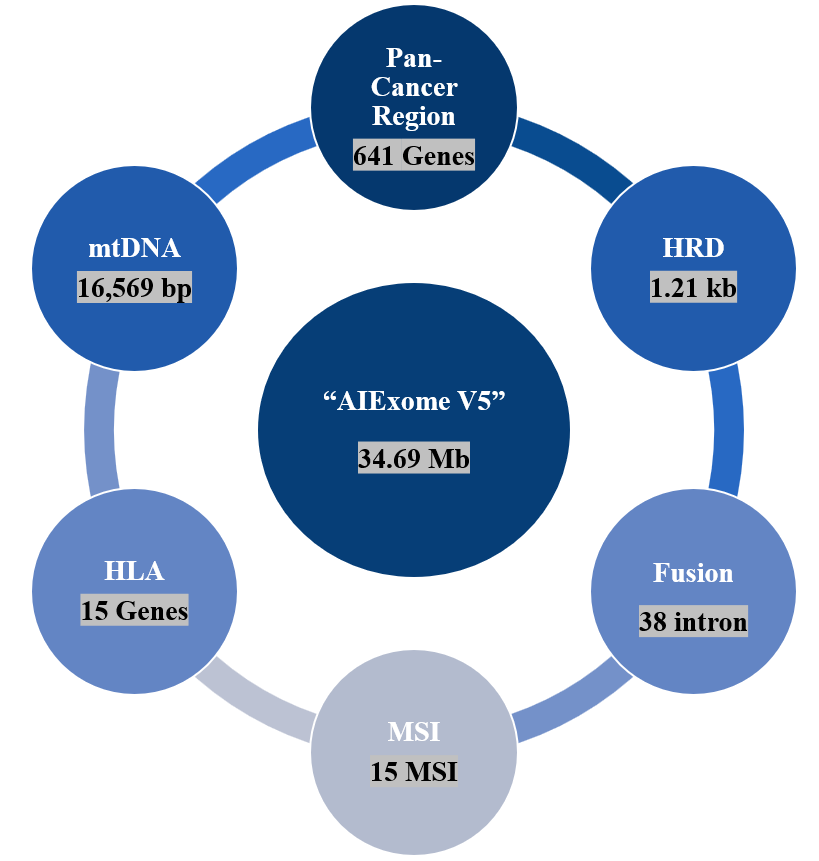
Figure 2. Coverage areas and marker coverage of AIExome V5-Tumor
Highlight 2: Effectively Save Up to 30G Data Volume
For routine somatic mutation detection of low-frequency mutations, a deduped depth of up to 500× is required for tumor genes to ensure detection accuracy. Conventional whole exome products on the market would require approximately 50 Gb of data for this purpose. However, the AIExome V5-Tumor features a unique depth adjustment technology that increases the depth of tumor-related regions by approximately 4 times compared to other exonic regions. Specifically, with 10 Gb of data and an average depth of 100× across the whole exome region, the depth for tumor-related genes can reach approximately 400×.
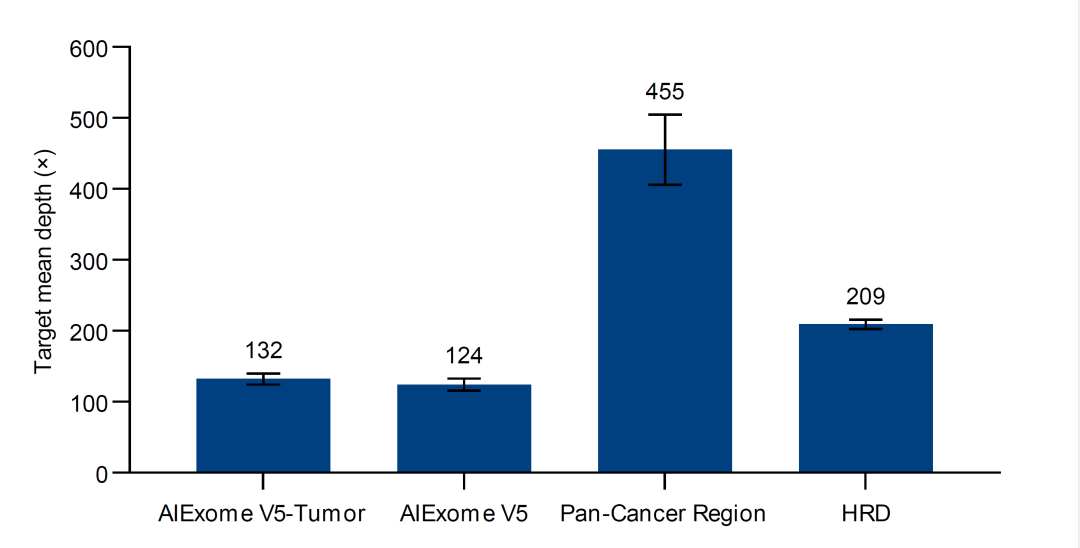
Figure 3. Depth ratio statistics of different probe panel regions in AIExome V5-Tumor
Highlight 3: Catering to Ultra-Fast Testing Cycle Requirements
Upgraded to a DNA probe-based whole exome version, it is more user-friendly and paired with 3.0 hybridization capture reagents, which can meet user needs with high standards for both overnight hybridization and rapid hybridization protocols.
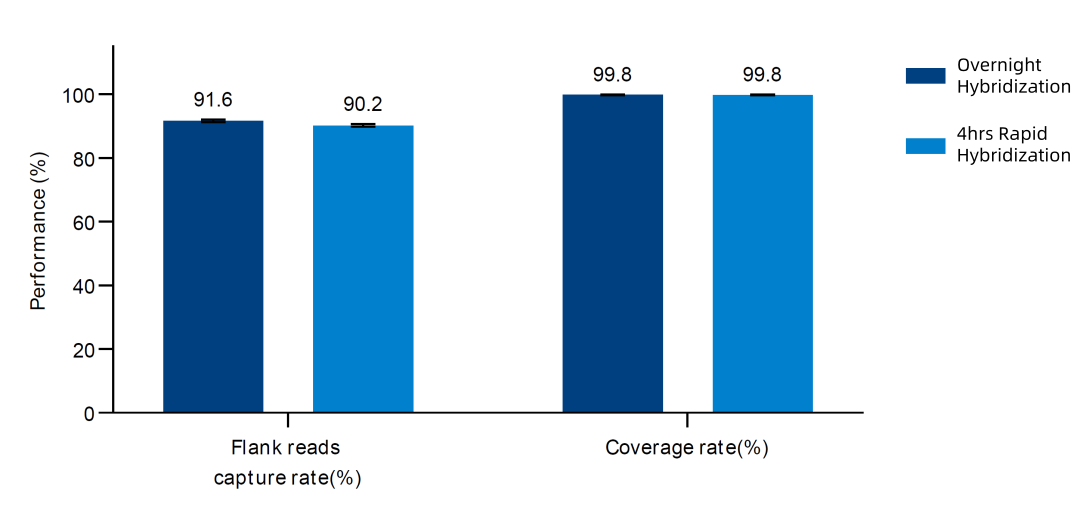
Figure 4. Capture performance of AIExome V5-Tumor probes paired with TargetSeq one v3.0 rapid hybridization and overnight hybridization.
Highlight 4: Accurate Detection of Variant Information
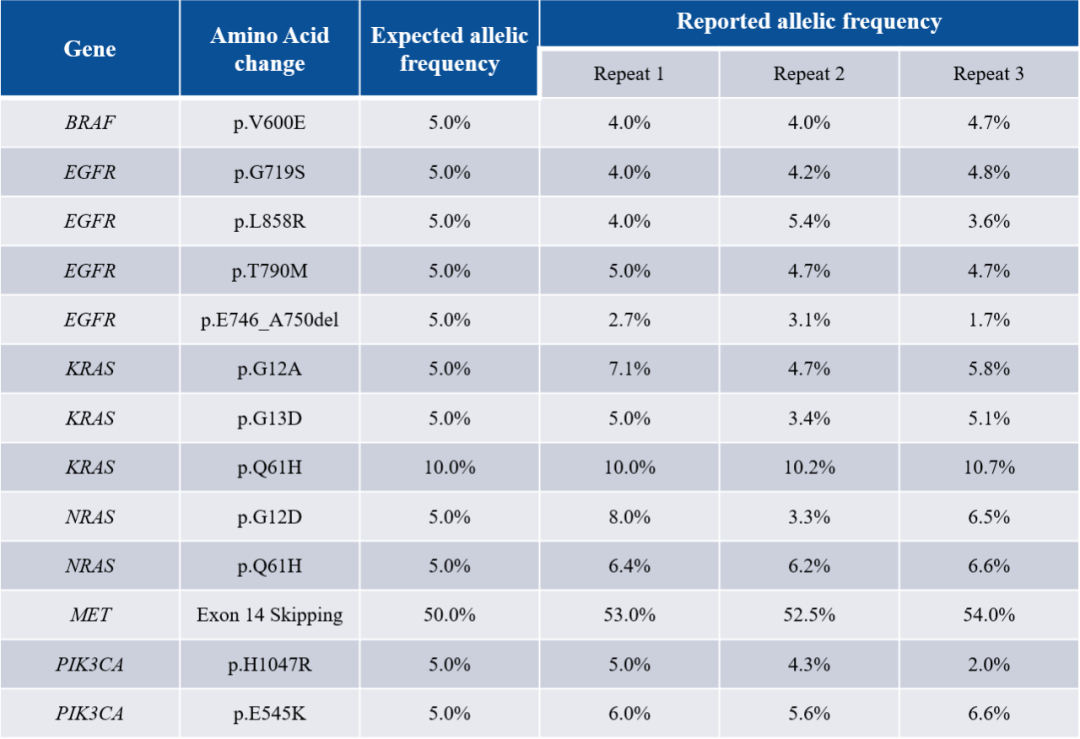
While saving data volume and increasing depth, the high-depth tumor-related regions enable accurate detection of low-frequency mutations.
Table 1. Mutation Detection Statistics in Positive Reference Materials
Supports Multiple Plus Solutions for Flexible and Convenient Customization of Intellectual Property-Exclusive Whole Exome Products
lIncludes numerous meticulously designed predefined product probe panels, ready-to-use after mixing, ensuring convenience and speed;
lEquipped with a 680K in-house developed probe synthesis platform to flexibly and rapidly address requirements for adding/encrypting regions;
lWith 11 years of experience in probe design, synthesis, and companion reagent product development, delivering more excellent and reliable product services.
AIExome V5-Tumor paired with Tumor-informed MRD free customization for comprehensive screening of monitoring sites enables sensitive and accurate MRD detection
In MRD (minimal residual disease) detection, the somatic sites screened vary by cancer type, with a median TMB (tumor mutational burden) of 3.6 mutations/Mb [1]. For this, using a ~2 Mb tumor large panel only screens approximately 7 sites, while ~35 Mb whole exome screening significantly increases the number to ~126 sites. This comprehensively covers patient-specific somatic mutation sites, significantly enhancing detection accuracy and sensitivity, and bringing better treatment plans and prognostic assessments for cancer patients.
Order Information

 CN
CN






