Plant Whole Exome Sequencing: Focus on Precise Gene Sequencing in Plants
Exons, as DNA regions directly involved in protein coding, carry the key genetic information of organisms. In plant research, Whole Exome Sequencing (WES) achieves precise sequencing of critical coding genes by target-enriching exonic regions in plant genomes. Take wheat as an example: in March 2025, researchers from Henan Agricultural University published a study in Nature Communications that used GWAS (Genome-Wide Association Study) and WES to identify TaCAT2, an enzyme associated with wheat stem base rot. The study revealed its interaction and phosphorylation with TaSnRK1α, ultimately proposing a resistance-mediated model of TaSnRK1α-TaCAT2 [1].
Relying on its independently developed TargetSeq® hybridization capture sequencing technology and IGT® Oligo Pools synthesis platform, iGeneTech has officially launched the innovative "Capture All" initiative. This plan aims to extend capture sequencing technology to broader life science fields, making it a foundational tool for multi-omics research and species evolution decoding. We will gradually launch hundreds of whole exome products, thousands of microbial whole genome capture products, multi-species methylome products, immune repertoire products, ancient DNA chips, and full-length mitochondrial capture probes for all known species.
Today, let's focus on plant whole exome products, enabling whole exome sequencing solutions for crops like wheat, barley, and maize to better support plant functional genomics and genetics research.
Perfect Integration of Technology and Application
While whole genome sequencing provides comprehensive data, it generates enormous datasets—especially for large genomes, which require even higher data volumes. For example, the wheat genome is approximately 14.6 Gb, and whole genome sequencing typically requires over 100 Gb of data, posing high analytical challenges and costs. In contrast, whole exome sequencing precisely focuses on exonic regions, enabling more efficient screening of genes associated with important plant traits. This saves researchers significant time and effort, driving the high-efficiency output of scientific achievements.
| WES | WGS |
Sequencing scope | Exonic regions | Whole genome |
Data volume | Lower | Higher |
Analysis complexity | Lower, focusing on exonic regions. | Higher, requiring comprehensive analysis of the whole genome. |
Storage space | Small | Large |
Cost | Lower | Higher |
Wheat whole exome product
Designed based on the 132.6 Mb coding DNA sequences (CDS) regions of the IWGSC RefSeq V2.1 reference genome, with over 2.55 million unique probes, it can efficiently and uniformly capture the CDS regions of over 100,000 wheat genes. A data volume of 5 Gb can achieve an effective depth of over 20×, suitable for scenarios such as wheat stress-resistant gene screening and variety evolution analysis.
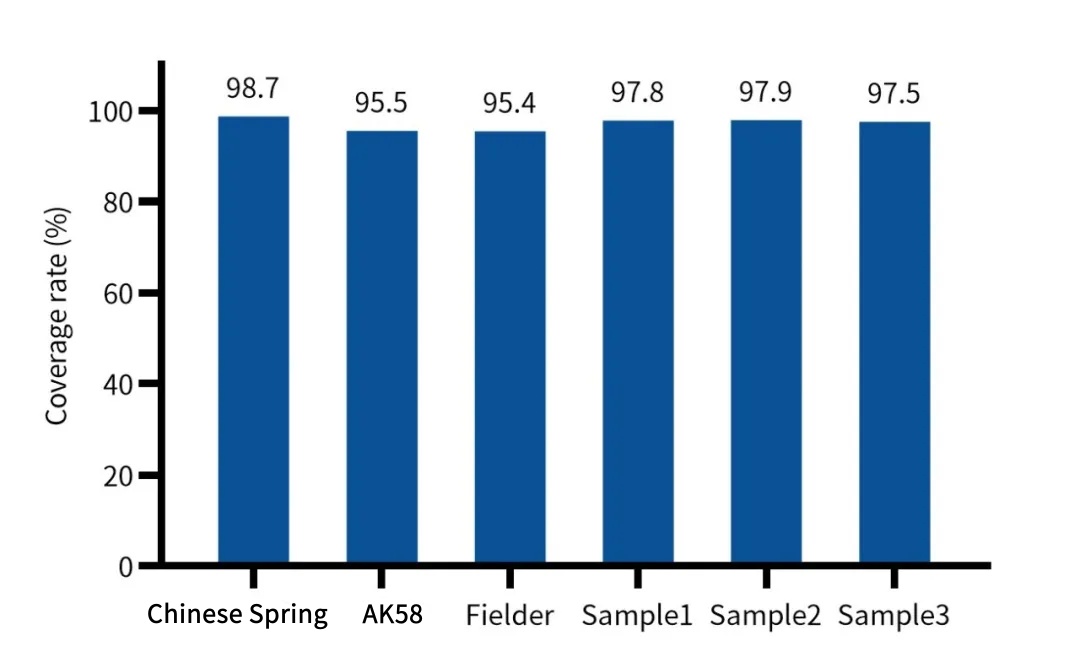
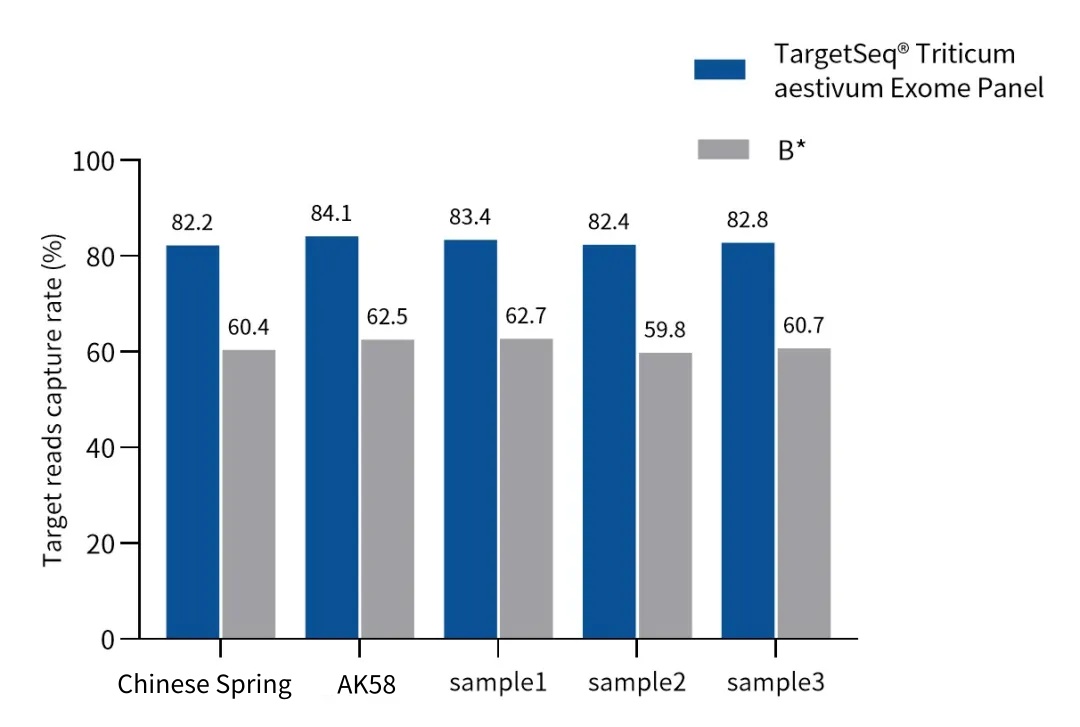

Figure 1. Capture performance of the Wheat Whole Exome Panel in different wheat varieties. A: Measured coverage of the CDS region, B: Capture efficiency, C: Uniformity.
Barley whole exome product
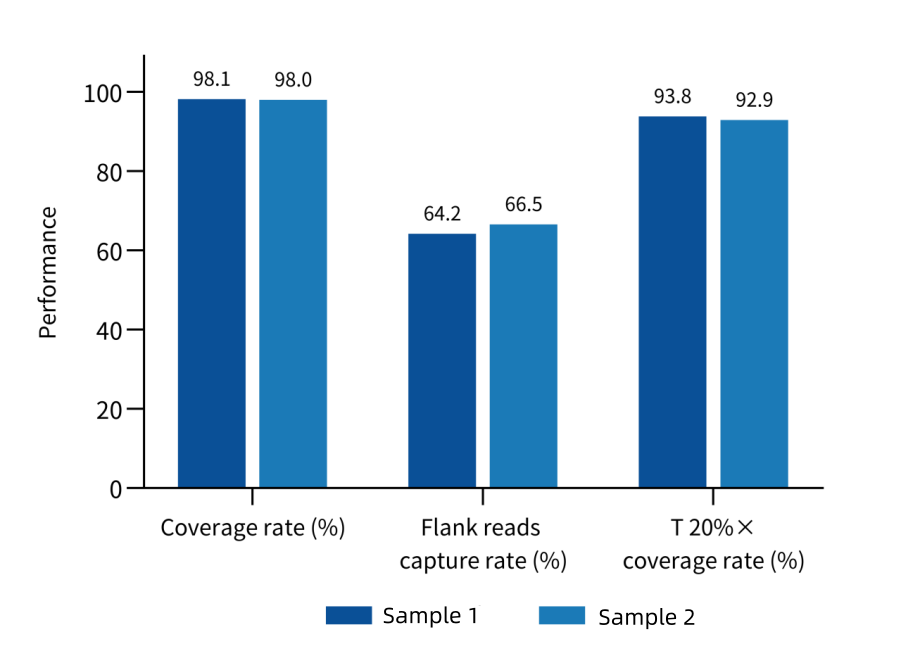
Designed based on the latest barley reference genome MorexV3, it covers 37.1 Mb of coding DNA sequences (CDS) regions, with over 460,000 unique probes, enabling capture of over 95% of the CDS regions of barley genes while combining high efficiency and uniformity. A data volume of 2 Gb can achieve an effective depth of over 20×, providing precise data for scenarios such as gene mapping for yield traits in barley and mutant library construction.
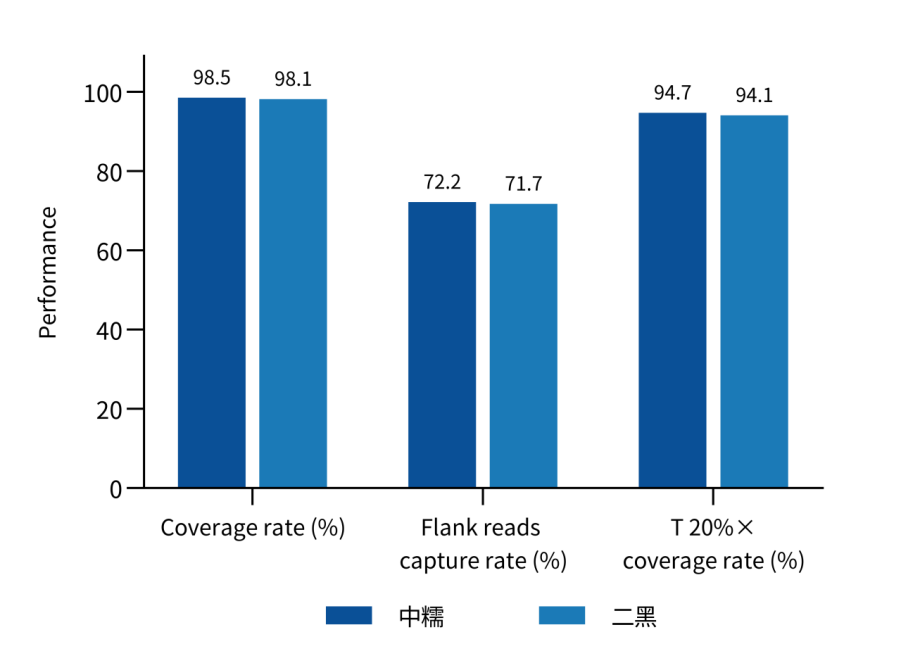
Figure 2. Capture performance of Barley Whole Exome Panel in different barley varieties.
Maize Whole Exome Product
Designed based on the latest maize reference genome B73 V5, it covers 43.2 Mb of coding DNA sequences (CDS) regions, with over 1.3 million unique probes, enabling efficient capture of over 96% of the CDS regions of maize genes. A data volume of 2 Gb can achieve an effective depth of approximately 20×, supporting cutting-edge research such as disease-resistant gene cloning in maize and analysis of heterosis mechanisms.
Order Information

 CN
CN





