In recent years, cfDNA-based liquid biopsy has been widely applied in oncology and other fields. However, due to the typically low yield of cfDNA extraction, library prep kits must meet extremely high performance requirements. The iGeneTech IGT™ Fast Library Prep Kit v2.0 is designed specifically for low-input cfDNA library preparation. With integrated UMI (Unique Molecular Identifier) technology, it supports the detection of low-frequency variants, making it suitable for ctDNA liquid biopsy and non-invasive prenatal testing (NIPT).
cfDNA and Its Applications
cfDNA (cell-free DNA), also known as circulating DNA, refers to free-floating DNA present in plasma, serum, or cerebrospinal fluid. ctDNA is a subtype of cfDNA derived from tumor cells, typically representing only a small fraction of total cfDNA. Hence, detecting cfDNA requires high-sensitivity technologies—NGS combined with UMI technology can reach sensitivities of approximately 0.03%. Today, cfDNA detection is widely used in early cancer screening, drug guidance, prognosis monitoring, prenatal screening, and microbial detection.
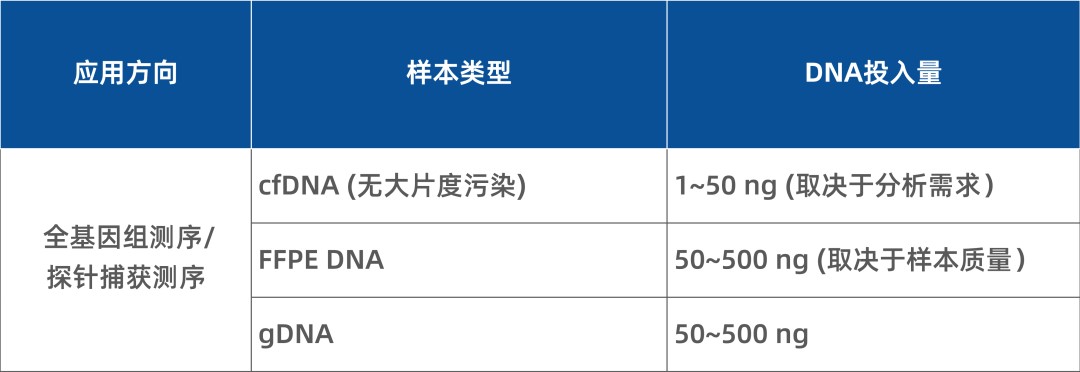
However, cfDNA levels in plasma are extremely low—typically only 1–30 ng per mL of plasma. The low proportion of ctDNA or fetal cfDNA further raises the demand for high molecular recovery and variant detection rates in cfDNA library prep kits.
Product Overview
Product Description
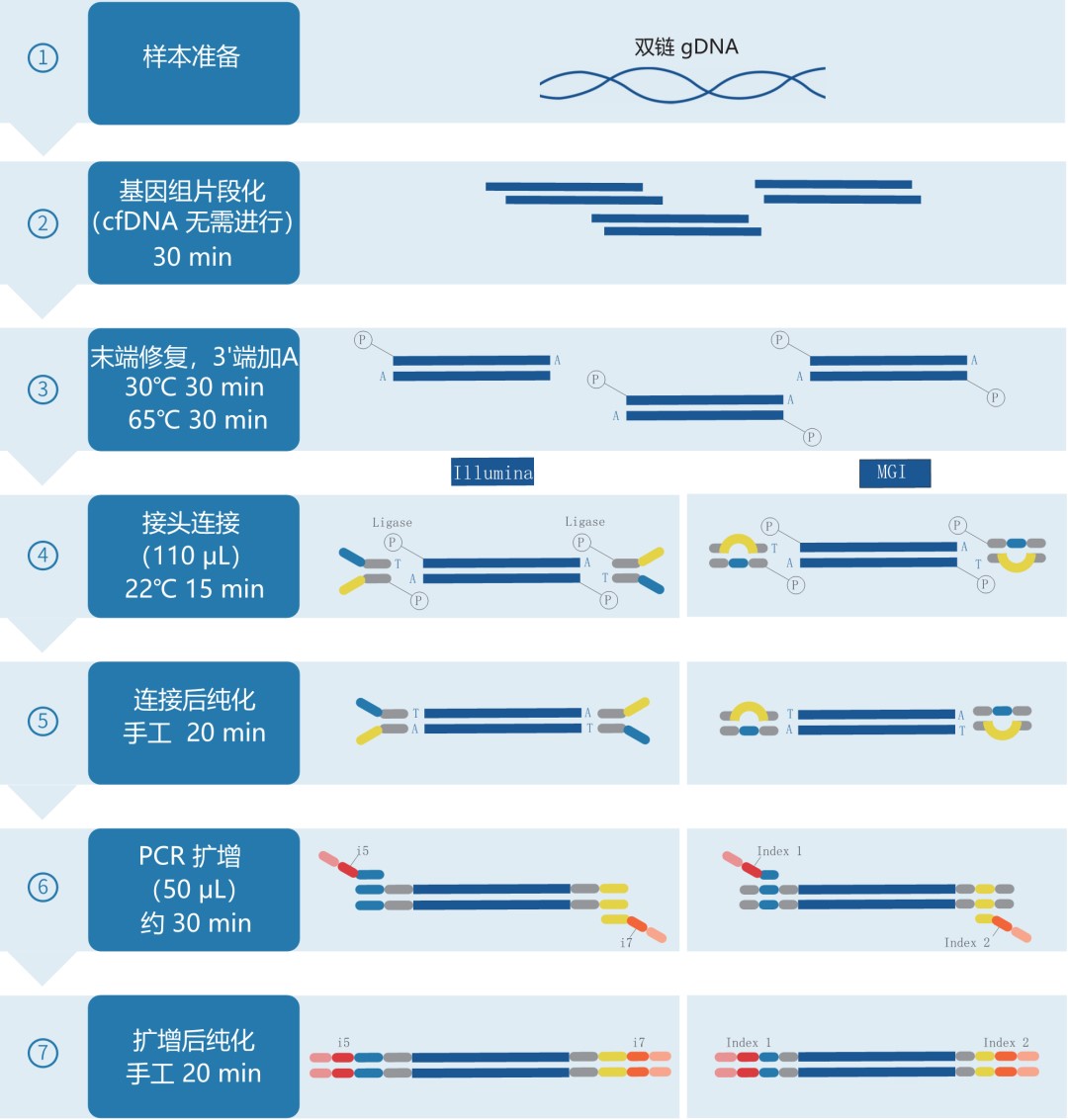
The IGT™ Fast Library Prep Kit v2.0 is a general-purpose double-stranded DNA library preparation kit suitable for cfDNA, FFPE DNA, and other samples. Based on A-T ligation chemistry, it features a simplified workflow and supports input amounts from 1 to 500 ng for both whole-genome sequencing and targeted capture sequencing. When used with appropriate adapters, it is compatible with Illumina and MGI sequencing platforms. UMI tagging enables sensitive detection of low-frequency variants.
Product Highlights
Low input requirement with high library conversion efficiency and consistent output
Compatible with cfDNA, FFPE DNA, gDNA, and more sample types
Supports UDI technology to reduce index hopping
Integrates UMI technology for low-frequency variant detection
Dual-platform compatibility: Illumina and MGI
Simple workflow, automation-friendly
Product Applications
Library Preparation Workflow
Product Advantages
Suitable for Low Input Library Construction
Samples such as cfDNA, biopsy tissues, and FFPE often have limited DNA yield. The IGT™ Fast Library Prep Kit v2.0 supports inputs as low as 1 ng. With inputs from 1–50 ng, final libraries exceeded 1500 ng. Under identical input and PCR conditions, this kit yields significantly higher library output than the K** kit and easily meets downstream capture requirements.
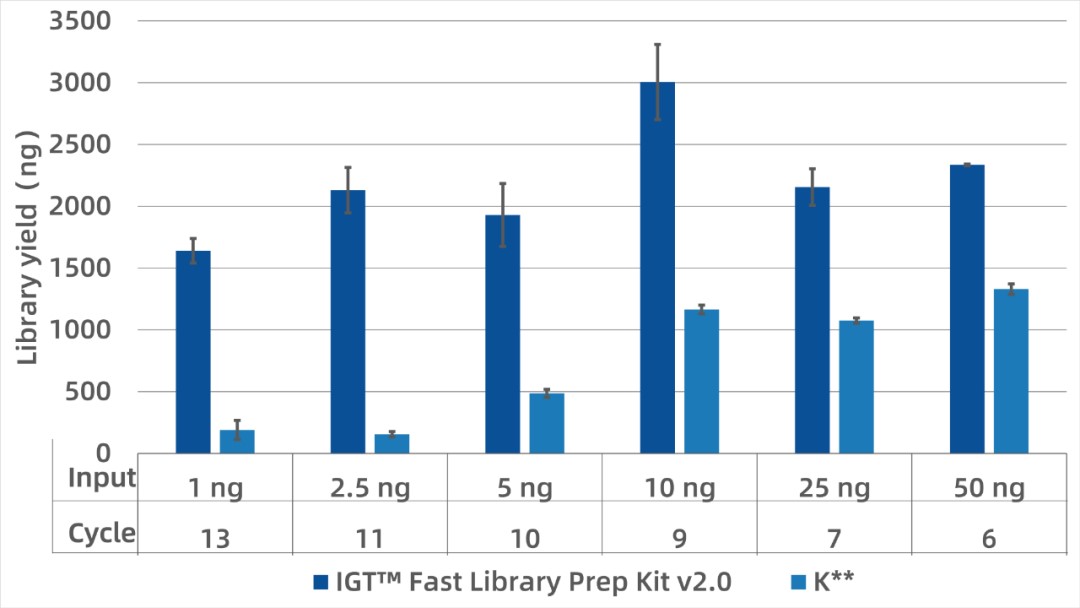
Figure 1. Library yield of IGT™ Fast Library Prep Kit v2.0 at various input levels.
gDNA was sonicated to 150–200 bp, and 1–50 ng was used with iGeneTech's Insert UMI Kit (for ILM). Amplification followed the recommended PCR cycles.
High Molecular Recovery Rate
10 ng of human cfDNA contains approximately 3,000 genome copies. A 0.1% variant would occur in only a few copies. This demands high recovery efficiency. When compared under equal input and sequencing depth (~20,000×), IGT™ Fast Library Prep Kit v2.0 produced 30–40% higher SSCS and DCS depths than the K** kit, indicating superior molecular recovery and conversion rates.
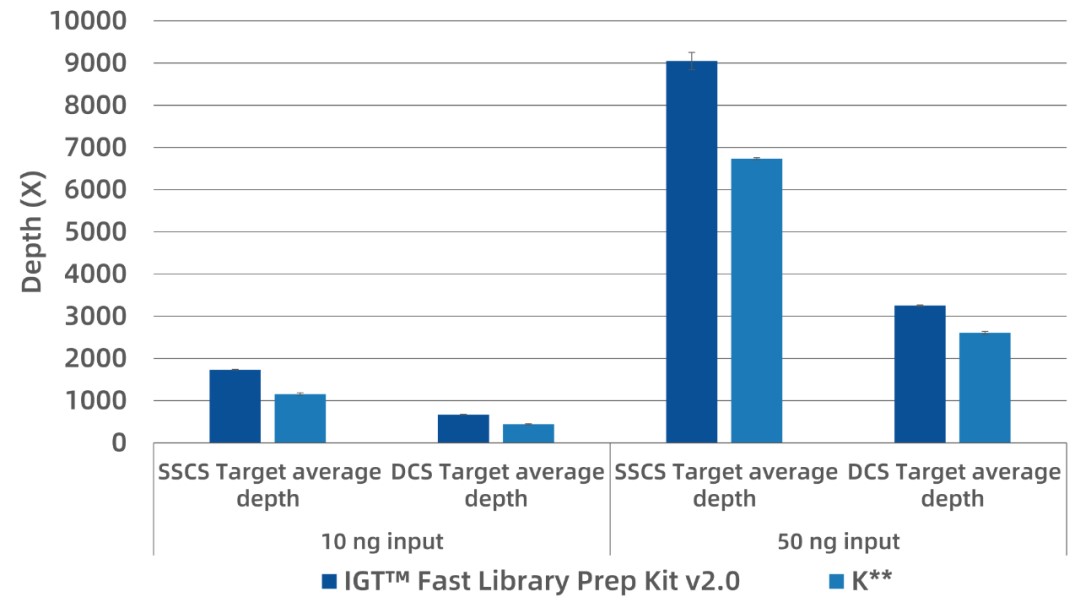
Figure 2. SSCS and DCS depth at ~20,000× pre-deduplication.
gDNA was sheared to 150–200 bp and inputs of 10 ng and 50 ng were used. Libraries were built with iGeneTech's UMI adapters, enriched using a 70 kb panel, and sequenced on Illumina NovaSeq 6000 (PE150). Analysis was done with iGeneTech's UMI pipeline.
High Variant Detection Consistency
Library prep and sequencing can introduce false positives, affecting low-frequency variant detection. When testing with 30 ng input and ~27,000× sequencing depth, intra-class correlation coefficient (ICC) for variants with VAF ≥ 0.5% was 1.000 between replicates, showing highly consistent results and minimized false positives.
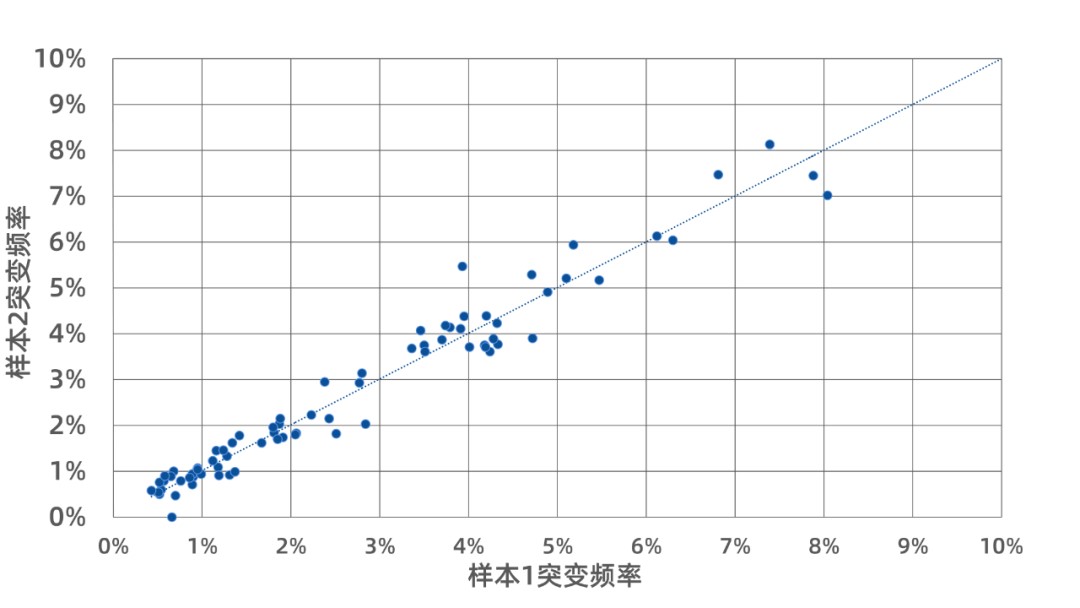
Figure 3. Variant consistency across replicates using IGT™ Fast Library Prep Kit v2.0.t v2.0.
Used 30 ng of 1% ctDNA standard (GeneWell, Cat. GW-OCTM009) with iGeneTech Insert UMI Kit and 70 kb capture panel. Sequencing and variant analysis performed on Illumina NovaSeq 6000.
High Accuracy in Low-Frequency Mutation Detection
A 0.5% cfDNA reference was made by mixing 1% mutation standard and wild-type standard (Horizon Discovery, Cat. HD780). Using 10 ng input with iGeneTech's UMI adapter, the library was enriched with a 70 kb panel and sequenced (~10,000× depth). All 8 known variants were detected with observed VAFs matching expected values. Sensitivity and specificity were both 100%.
Table 1. Detection of 0.5% known mutations using IGT™ Fast Library Prep Kit v2.0.
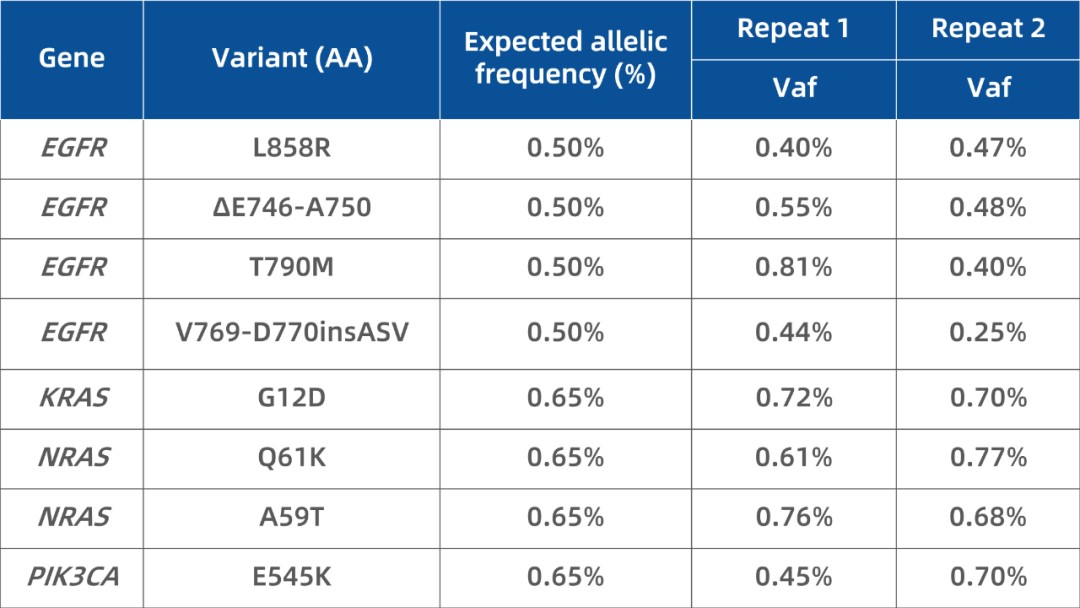
(*Specificity based on non-variant base calls across designated genomic loci.)
Product Information
About iGeneTech®
iGeneTech is a national high-tech enterprise in China, dedicated to gene capture technologies. The company owns independently developed hybridization probe and multiplex PCR gene capture platforms. With large-scale diagnostic service capabilities, iGeneTech provides world-leading, full-cycle gene capture technology solutions.
After years of industrial development, iGeneTech now offers a complete portfolio of upstream to downstream gene capture solutions, including:
Custom gene capture product development
LIMS systems for labs
Fully automated laboratory workflows
Data management and interpretation solutions
Laboratory quality management systems
Ultra-fast NGS sequencing services
Ultra-high-throughput sample processing diagnostics
As a global provider of end-to-end sequencing solutions, iGeneTech partners with third-party clinical labs, precision medicine centers, genetic testing companies, and clinical researchers, delivering highly automated, intelligent, productized ALL-IN-ONE gene testing solutions.

 CN
CN