Spinal Muscular Atrophy (SMA)
Spinal muscular atrophy (SMA) is a hereditary neuromuscular disorder categorized into 5q-SMA (caused by mutations in the SMN1 gene) and non-5q-SMA (resulting from mutations in genes such as IGHMBP2, SCO2, etc.). However, due to the genetic heterogeneity of SMA and the diversity of clinical manifestations, precise diagnosis faces enormous challenges.
Traditional methods such as MLPA (multiplex ligation-dependent probe amplification) and Sanger sequencing can detect SMN1 gene deletions or point mutations, but they have limited ability to analyze non-5q-SMA and complex genotypes.
In the study "Unraveling the genetic mysteries of spinal muscular atrophy in Chinese families"【1】, researchers systematically analyzed the genetic characteristics of 680 Chinese SMA families by integrating multiple molecular techniques, particularly whole exome sequencing (WES). The study revealed the core value of WES in enhancing diagnostic efficiency.
Research Methods and Technical Innovations
The research team adopted a multi-technology combined strategy:
lMLPA and Long Range PCR: Rapid screening of copy number variations and common deletions in the SMN1 gene.
lSanger Sequencing: Validation of specific point mutations (e.g., c.840C>T).
lWhole Exome Sequencing (WES): Using the AIExome V2 Genetic Enhanced Probe (covering all exonic regions with 100% coverage of pathogenic sites in key genetic disease databases), WES was applied to identify rare gene mutations, resolve complex cases, and correct false-positive results from other techniques.
Advantages of WES Technology
lComprehensiveness: Detect over 20,000 genes in a single run without pre-assuming candidate genes.
lHigh Resolution: Identification of single-nucleotide variants (SNVs), small insertions/deletions (Indels), and complex structural variations.
lDeep Coverage: Average sequencing depth>100×, ensuring detection of low-abundance variants.
Core Findings: The Critical Role of WES
1. Revealing Rare Pathogenic Genes in Non-5q-SMA
Among 680 families, 3 carried IGHMBP2 gene mutations (causing SMARD1 and CMT2S), 1 carried a SCO2 gene mutation (mitochondrial disease), and 1 carried a NEB gene mutation (nemaline myopathy). All pathogenic genes in these non-5q-SMA cases were identified via WES. Traditional methods failed to detect such rare mutations due to their targeted limitations, establishing WES as the gold standard for non-5q-SMA diagnosis.
2. Correcting MLPA False Positives to Avoid Misdiagnosis
The study reported 2 cases of MLPA false positives:
Case 1: MLPA showed a homozygous deletion of the SMN1 gene in a child, but WES revealed compound heterozygous mutations in the NEB gene (R8358Vfs20 and E8167), leading to a final diagnosis of nemaline myopathy.
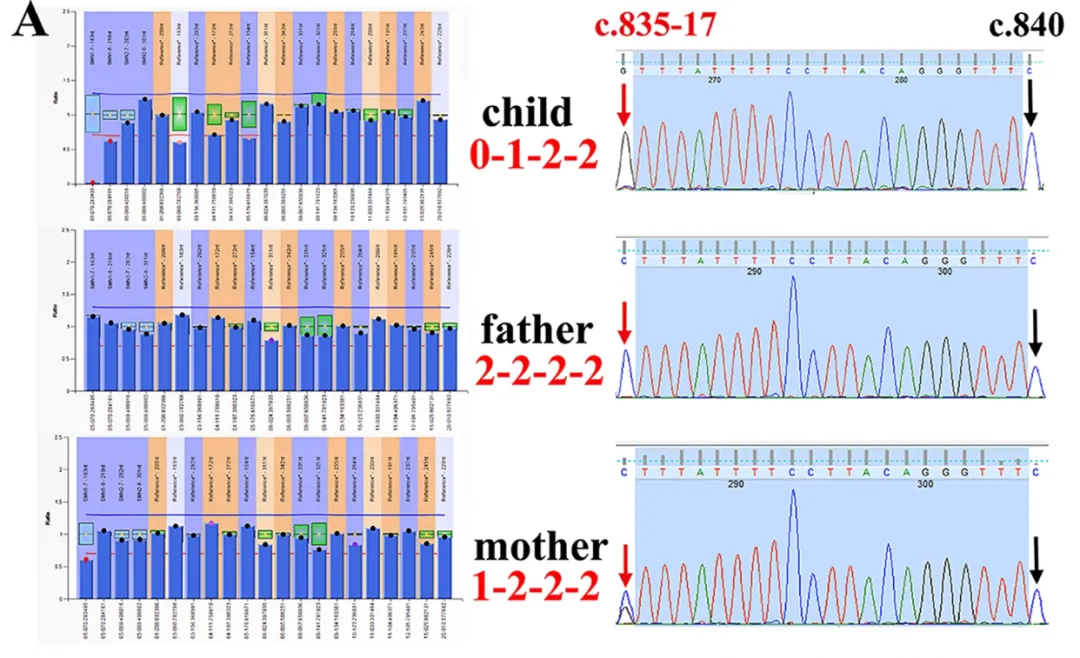
Figure 1. MLPA false positive result 1.
The MLPA result indicated a homozygous deletion of exon 7 (E7) of the SMN1 gene in the child, with a preliminary diagnosis of SMA. The mother was identified as a "1+0" carrier (one normal copy + one deleted copy), and the father was inferred to be a "2+0" carrier (two normal copies + one deleted copy).
Case 2: MLPA indicated a single copy of SMN1 in the father, but WES and digital PCR (dPCR) confirmed a double copy. Combined with Sanger sequencing, a pathogenic point mutation (c.835-5 T>G) in the SMN1 gene was identified.
Through comprehensive gene screening, WES effectively avoided false positives caused by mutations in the MLPA probe binding region, significantly enhancing diagnostic accuracy.
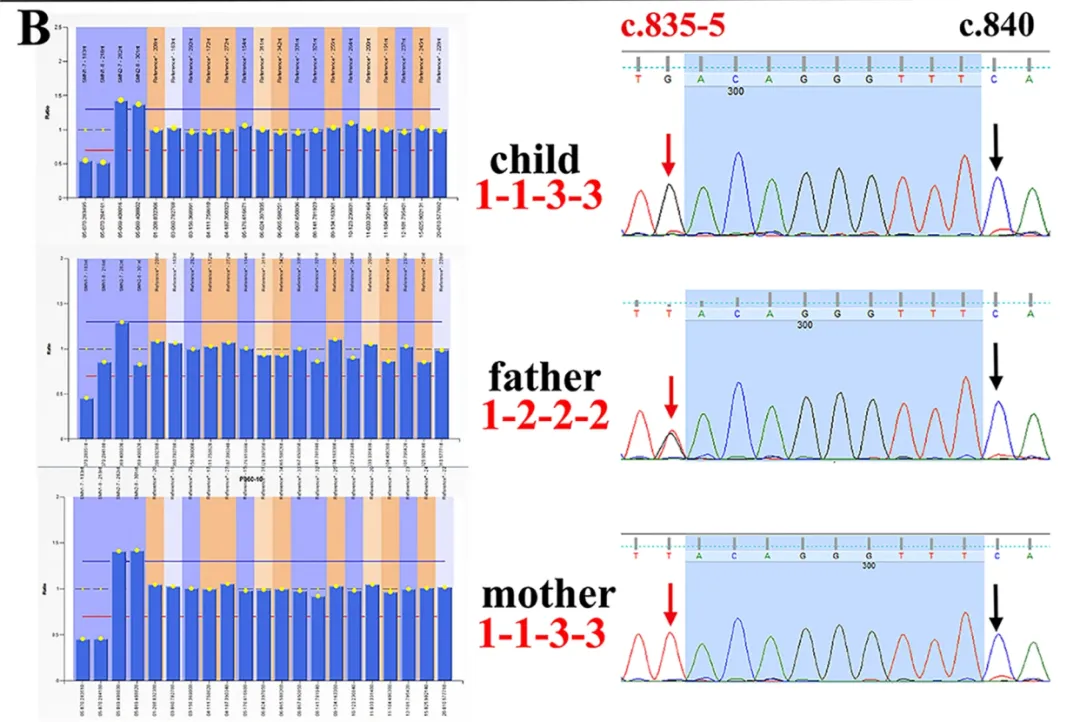
Figure 2. MLPA false positive result 2. The father’s MLPA result showed only 1 copy of SMN1 E7 (exon 7), but both dPCR (digital PCR) and sequencing indicated he had two copies of SMN1 E7. Further sequencing confirmed he carried a pathogenic variant in the SMN1 gene: c.835-5 T>G.
3. Deciphering the Genetic Mechanisms in Asymptomatic Patients
Five asymptomatic individuals carried homozygous deletions of the SMN1 gene (e.g., a 40-year-old pregnant woman in Family A), but their clinical manifestations did not align with their genotypes. WES analysis revealed these individuals had heterozygosity in the SMN1/SMN2 genes (e.g., c.*239A>G variants) or carried other modifier genes (e.g., PLS3 overexpression), explaining the phenotypic heterogeneity. WES provided a molecular basis for genetic counseling in asymptomatic carriers.
4. Optimizing the Clinical Diagnostic Workflow
The study proposed a WES-integrated SMA diagnostic pathway:
lFor MLPA-negative but clinically highly suspected cases, prioritize WES to screen for non-5q-SMA.
lFor MLPA false positives or complex genotypes (e.g., "2+0" carriers), use WES to validate and identify true pathogenic mutations.
lThe integration of WES shortened the diagnostic cycle and reduced the risk of missed or incorrect diagnoses.
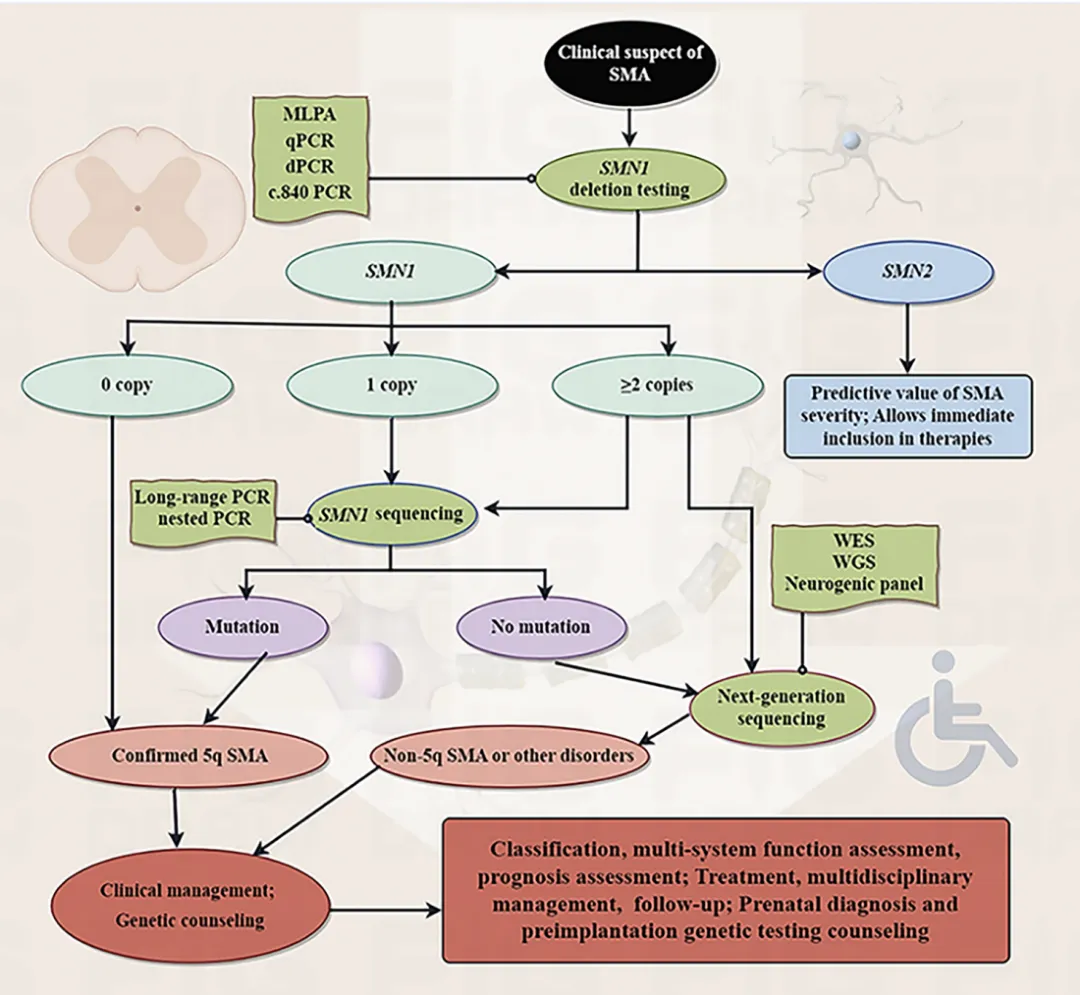
Figure 3. Flow Chart of SMA Genetic Testing and Genetic Counseling
Conclusion
Through the analysis of a large-scale Chinese SMA family cohort, this study confirmed the core role of whole exome sequencing (WES) in the following aspects:
lPrecisely identifying rare pathogenic genes in non-5q-SMA and expanding the disease mutation spectrum.
lCorrecting false-positive/false-negative results from traditional methods to enhance diagnostic reliability.
lDeciphering complex cases with phenotype-genotype discordance to advance personalized medicine.
The integration of WES not only optimizes the clinical diagnostic pathway for SMA but also provides a paradigm for molecular mechanism research in other hereditary diseases. In the future, combining long-read sequencing and functional validation will further enhance the application potential of WES.
This study thoroughly analyzed the genetic characteristics of suspected familial SMA cases in the Chinese population through in-depth research on the SMN1 gene, revealed the molecular genetic mechanisms of SMA, and provided a critical basis for clinical genetic counseling, prenatal diagnosis, and targeted therapy.
WES technology plays an irreplaceable role in detecting non-5q-SMA cases, correcting detection errors, and exploring genetic variants. With the popularization of WES and broader genomic sequencing technologies, molecular diagnostics for SMA and other genetic disorders will become more comprehensive and efficient.
Related Products
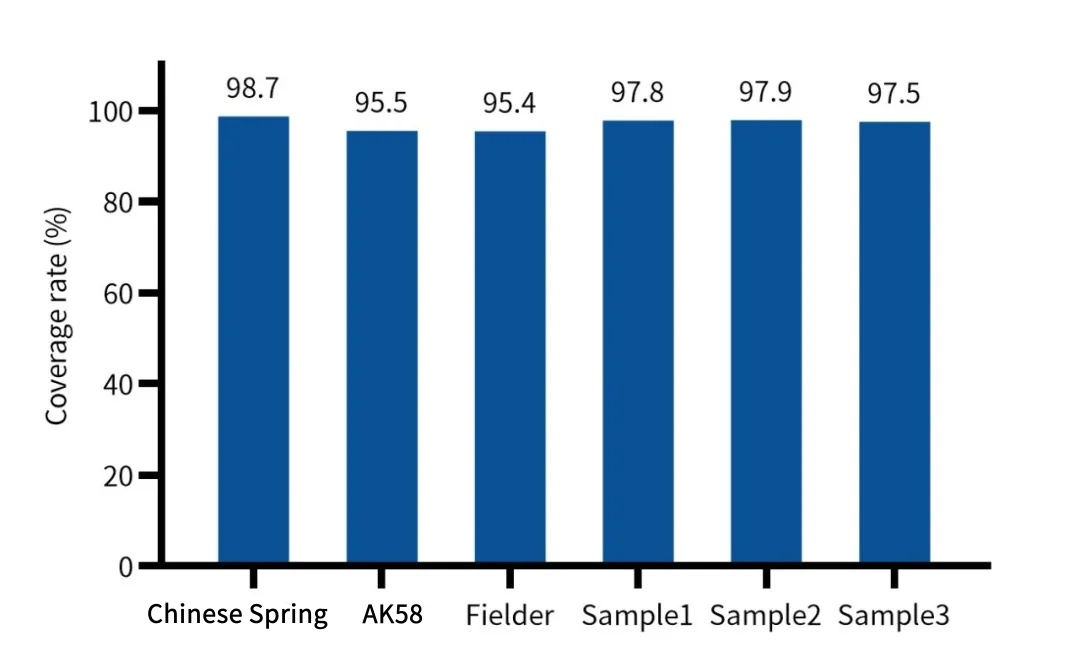
AIExome V5: Redefining Industry Standards with Exceptional Uniformity and High-Efficiency Capture Technology
Core Feature 1: Exceptional Uniformity
Innovative probe design significantly reduces GC bias, with a Fold 80 value as low as 1.24—consistently outperforming market products.
Core Feature 2: Ultra-Fast and Efficient Capture
Minimum 2-hour rapid hybridization protocol efficiently achieves high-quality targeted fragment capture, balancing speed and data performance.
Core Feature 3: Comprehensive and Precise Coverage
Intelligent overlapping probe design increases 20×/30× coverage ratio, particularly optimizing difficult-to-cover regions of OMIM genes.
References
【1】Gao S, Chen D, et al. Unraveling the genetic mysteries of spinal muscular atrophy in Chinese families. Orphanet J Rare Dis. 2025 Jan 15;20(1):25. doi: 10.1186/s13023-024-03523-0.

 CN
CN