Whether the mutation results obtained from different WES sequencing protocols and data analysis methods are reliable and consistent has become a key concern for researchers and clinicians.
In the R&D-to-production transition phase, performance validation of whole exome sequencing (WES) is inseparable from consistency analysis. By precisely evaluating the accuracy and reliability of data, it lays a solid foundation for the smooth transition of research outcomes to clinical applications. In the production quality control process, regular data verification and cross-contamination screening rely on consistency analysis as a "sharp eye" to monitor the production process in real time, ensuring the purity and authenticity of every piece of sequencing data.
Since the release of AIExome V5 Core, Inherit, and Tumor Edition, their technical indicators in wet experiments have performed excellently, gaining extensive attention and application.
Next, through a series of thematic articles, we will deeply dissect the practical pathways and cutting-edge value of consistency analysis for next generation sequencing (NGS) mutation results.
In this first chapter, we will systematically analyze the consistency evaluation methods between WES data and NA12878 standard mutation results.
Background: What is NA12878?
GIAB (Genome in a Bottle) is an initiative launched by the U.S. National Institute of Standards and Technology (NIST). Its goal is to create high-quality, widely recognized reference datasets of the human genome for evaluating the performance of genome assembly and analysis tools. The NA12878 sample is derived from a healthy female of Northern and Western European ancestry. With a reliable source, it is managed by the authoritative Coriell Institute for Medical Research (USA) and has been extensively used and validated worldwide, serving as a "benchmark" in genomics research. GIAB has conducted in-depth sequencing and analysis of NA12878, designating it as HG001. As the standard variant dataset for NA12878, HG001’s mutation information has been validated through multiple high-precision methods, and it is frequently used for benchmarking and method validation in genomics research.
In whole exome sequencing studies, consistency analysis between mutation results derived from NA12878 sequencing data and its known mutation results enables the evaluation of WES technology’s accuracy in detecting gene mutations. Such analysis is often applied to assess the performance of exome sequencing products, sequencing platforms, and data analysis pipelines, ensuring the robustness and reliability of subsequent data.
Method: How to Obtain NA12878 Data
NA12878 variant dataset and high-confidence regions: The GIAB project has constructed an authoritative set of mutation detection and high-confidence regions by integrating sequencing data from multiple technical platforms. As an industry-recognized "gold standard" for validating variant detection workflows, it provides a core reference for evaluating detection accuracy and consistency. The relevant variant set VCF files and supporting confident region BED files can be obtained from the following address: https://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/release/NA12878_HG001/
Analysis Steps
1. Obtain the NA12878 standard sample variant dataset and its high-confidence regions, and calculate the intersection region between these regions and the whole exome target regions.
2. Extract the NA12878 variant results and whole exome standard variant results within this intersection region.
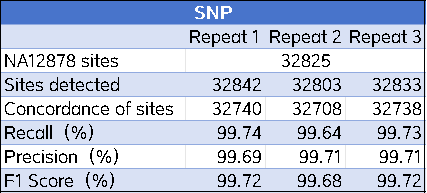
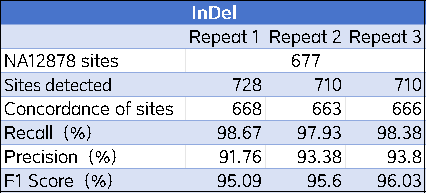
3. Evaluate the numbers of TP (True Positives), FP (False Positives), and FN (False Negatives) among the SNP and InDel loci included in the intersection region, thereby calculating indicators such as Recall and F1 Score.
Calculation Formulas
NA12878 |
Test sample | Predicted mutation, actual mutation (True Positive, TP) | Predicted mutation, actual no mutation (False Positive, FP) |
Predicted no mutation, actual mutation (False Negative, FN) | Predicted no mutation, actual no mutation (True Negative, TN) |
Precision: TP/(TP+FP)
Sensitivity: Also known as Recall or True Positive Rate (TPR), it is a key indicator for evaluating the detection capability of a model. It represents the proportion of samples that are truly positive and correctly identified as positive by the model among all truly positive samples.
The calculation formula is: TP/(TP+FN).
F1 Score is a comprehensive indicator used to evaluate the performance of classification models in machine learning and statistics, especially suitable for scenarios with imbalanced positive and negative samples. Its significance lies in balancing the trade-off between Precision and Recall, resolving potential contradictions between them in practical applications, and avoiding the one-sidedness of a single indicator.
The formula is as follows:
F1 Score = 2 × (Precision × Recall) / (Precision + Recall)
The performance of the AIExome V5-Inherit product in detection was evaluated using the NA12878 standard dataset. The results showed that this product performed excellently in SNP and Indel detection. For SNP detection, the Recall reached 99.74%, the Precision reached 99.71%, and the F1 Score exceeded 99.72%; the F1 Score for Indel detection was also higher than 96%, demonstrating the high reliability and accuracy of this product in clinical and research applications.

 CN
CN



