There is currently a lack of a unified standard for how to choose the Minimal Residual Disease (MRD) detection strategy, and there are still some doubts about its clinical application. In this article, we will provide relevant reference information for you regarding the current situation of the application of MRD detection strategies.
ctDNA MRD Detection Strategy
The detection strategies based on ctDNA MRD mainly fall into the following two categories [1]:
Tumor-agnostic assays
Tumor-agnostic analysis, which is independent of the primary tumor tissue. It detects and analyzes ctDNA based on a pre-selected fixed panel of tumor-related genes. In addition to analyzing ctDNA mutations, it often uses multi-omics methods such as the combination of ctDNA methylation or fragmentomics to assist in monitoring Minimal Residual Disease (MRD), providing a more comprehensive reference for auxiliary diagnosis and medication.
Tumor-informed assays
Tumor-informed analysis. In this scheme, a comprehensive genomic sequencing (Whole Exome Sequencing, WES, or a fixed large panel) is first performed on the primary tumor tissue to obtain the patient's genomic variation profile. Then, a certain number of known mutation sites are selected to customize a personalized panel for subsequent dynamic detection.
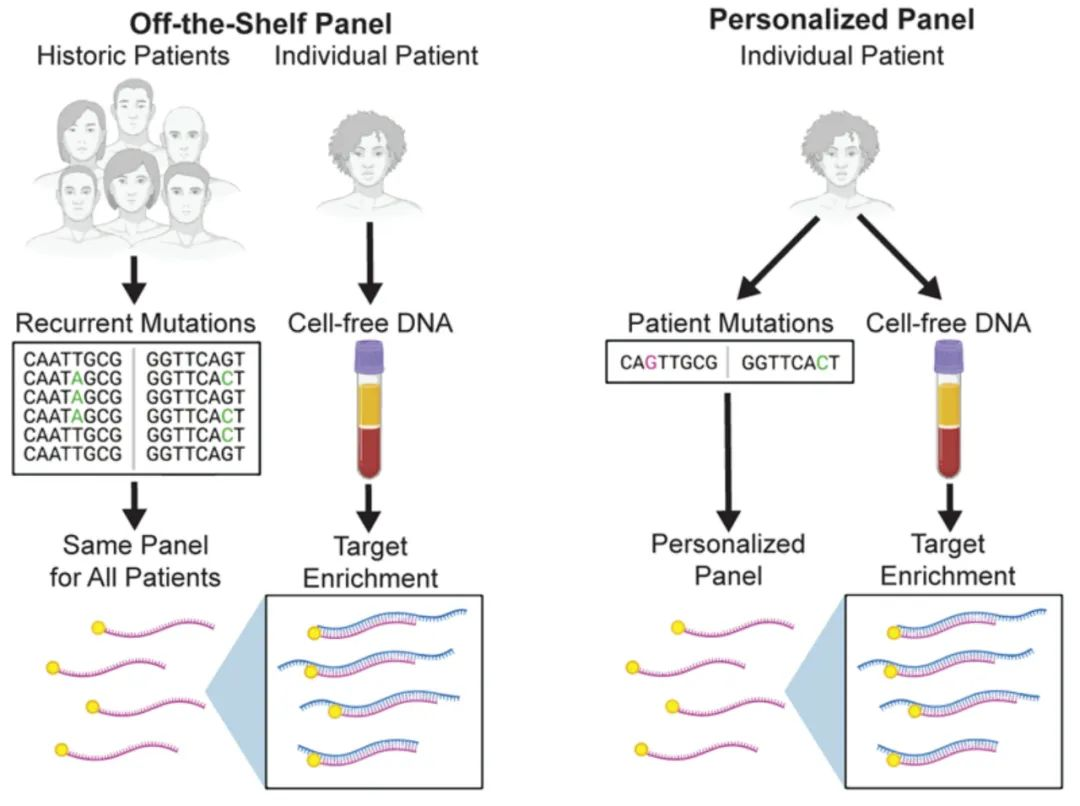
Figure 1. Comparison between the personalized panel for patients with Minimal Residual Disease (MRD) in solid tumors and the cancer-type fixed panel [2]
Different technical routes have their own advantages and disadvantages in clinical applications. Currently, there is no clear conclusion on which method is more superior. Previous literature reports show that these two MRD detection technologies are very close in detection performance. A mature personalized panel for patients and a fixed panel are comparable in various aspects, such as the detection rate of ctDNA after lung cancer surgery and the performance of MRD prognosis monitoring [3,4].
Domestic Layout of MRD Detection Strategies
Currently, the MRD detection products launched by domestic tumor gene testing companies are also diverse. In addition to the basic solutions of tumor-informed assays and tumor-agnostic assays, some testing companies have also integrated the two strategies. However, from the overall distribution, tumor-informed assays occupy the main position.
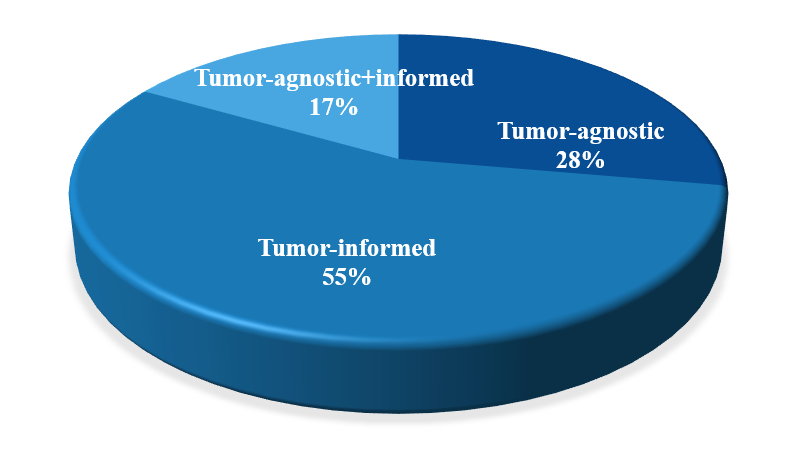
Figure 2. The Proportion of Strategies of Different Domestic MRD Detection Products Currently
On March 31, 2024, at the 13th Pathology Annual Meeting, Professor Wu Huanwen from Peking Union Medical College Hospital delivered a report titled "Consensus on the Detection of Molecular Residual Disease (MRD) in Solid Tumors". Regarding the ctDNA MRD detection strategy, it was also mentioned that the tumor-informed analysis strategy is recommended, and the relevant consensus content is as follows:
Consensus 3.3
"The tumor-informed analysis based on tumor tissue mutations has better sensitivity and specificity compared to the tumor-agnostic analysis; The tumor-informed analysis strategy should be given priority."
Consensus 3.4
When adopting the tumor-informed analysis strategy, whole exome sequencing (WES) can be used for tumor tissue detection, or a multi-gene panel for solid tumors can also be selected. WES has a comprehensive coverage range, but its analysis complexity is high. The multi-gene panel is convenient to implement, but its detection range is limited. In clinical practice, comprehensive considerations should be taken for the selection.
Why Choose Tumor-informed Assays?
⊙ In the CIRCULATE-Japan study, a tumor-informed customized panel based on WES was used. A total of 8,374 genes were selected for monitoring 1,039 patients, among which the proportion of unique genes for each patient was as high as 55.7% [5].
⊙ At the 2022 WCLC meeting, a retrospective analysis study was released. A tumor-informed customized panel based on WES was used. 4,549 sites were selected for MRD detection in 163 lung cancer patients, covering 3,343 genes. Among them, 81.5% of the genes were unique to a single patient, and the clinical significance of 94.1% of the gene variations was not yet clear, indicating that these genes were probably not within the scope of conventional panel design [6].
⊙ At the 2023 ASCO GI, a clinical study on MRD in colorectal cancer was released. A tumor - informed customized panel was used. 5,835 sites were selected for 117 patients to design a customized panel. Only 6% of the variant sites could be covered by the fixed panel, and 75% of the genes were unique to each patient [7].
⊙ In 2023, the Cancer Cell published the results of the MEDAL (NCT03634826), the world's first prospective clinical study that head-to-head compared three MRD detection strategies in early-stage lung cancer. According to the pre-operative blood test results, the detection sensitivity of the tumor-informed customized panel strategy was better than that of the tumor - agnostic fixed panel strategy, and the tumor-informed customized panel could better predict the disease-free survival (DFS) of patients [8].
From the above research results, it can be found that during the process of MRD monitoring, the uniqueness of the molecular characteristics of each patient's tumor needs to be fully considered. Compared with the fixed panel, the individualized MRD detection scheme selects personalized monitoring sites for each patient. Therefore, it can fully ensure the accuracy and sensitivity of MRD monitoring.
Selection of Individualized MRD Detection Sites
In the tumor-informed detection strategy, the number of monitoring sites selected for each patient among the patient-specific sites obtained from WES detection is also crucial for the sensitivity of subsequent MRD detection. In a study on the detection performance of ctDNA MRD, a panel with 165 sites was used to conduct a mixing test on cell lines with known mutations and normal cell lines at ratios ranging from 1% to 0.001%. At a ratio of 0.001%, two mutations could still be detected. Moreover, in a simulation experiment where 10-150 sites were randomly selected for detection, it was found that when the mixing ratio was relatively high (≥0.5%), the sensitivity was not affected by the number of detection sites. However, when the mixing ratio was less than 0.01%, the sensitivity increased significantly with the increase in the number of detection sites [9]. Therefore, in ctDNA samples with low abundance, the detection sensitivity can be improved by increasing the number of sites in individualized MRD detection.
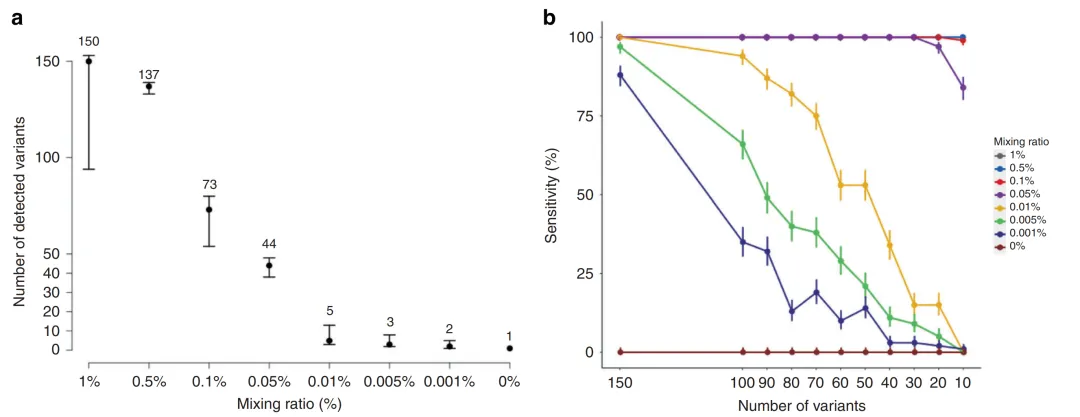
Figure 3. The Number of Detection Sites and Detection Sensitivity [9]
The Free Customization Plan of iGeneTech's Individualized MRD Panel
During the current promotion of MRD detection, individualized customization clearly has distinct advantages. Increasing the number of detection sites can significantly boost detection sensitivity. However, the customization costs resulting from more sites are a major limiting factor.
To address this, iGeneTech will roll out a free customization plan for an individualized MRD panel with up to 150 sites. Meanwhile, it offers an integrated solution covering the whole process, including extraction, library preparation, capture, and UMI analysis. We aim to work with all stakeholders to expedite the maturation of MRD detection technology and its clinical application, benefiting more patients.
References
1. Wu Yilong, Lu Shun, Cheng Ying, et al. Expert Consensus on Molecular Residual Disease in Non-Small Cell Lung Cancer. Evidence-Based Medicine, 2021,21(3): 129-135.
2. Everett J, Moding., Barzin Y, Nabet., Ash A, Alizadeh., Maximilian, Diehn.(2021). Cancer Discov, 11(12), 0. doi:10.1158/2159-8290.CD-21-0634
3. Aadel A Chaudhuri, Jacob J Chabon, Alexander F Lovejoy, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017 Dec;7 (12):1394-1403. doi: 10.1158/2159-8290.CD-17-0716.
4. Christopher Abbosh, Nicolai J Birkbak, Gareth A Wilson, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017 Apr 26;545 (7655):446-451. doi: 10.1038/nature22364
5. Kotani D, Oki E, Nakamura Y, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med. 2023;29(1):127-134.
6. https://doi.org/10.1016/j.annonc.2023.09.1749
7. https://ascopubs.org/doi/abs/10.1200/JCO.2023.41.4_suppl.213
8. Chen K, Yang F, Shen H, et al. Individualized tumor-informed circulating tumor DNA analysis for postoperative monitoring of non-small cell lung cancer. Cancer Cell. 2023;41(10):1749-1762.e6.
9. Seung-Bum, Ryoo,Sunghoon, Heo,Yoojoo, Lim et al. Personalised circulating tumour DNA assay with large-scale mutation coverage for sensitive minimal residual disease detection in colorectal cancer.[J] .Br J Cancer, 2023, 129: 0.

 CN
CN



