In RNA-seq transcriptome sequencing research, as the primary source of interference, rRNA (ribosomal RNA) accounts for 80%-90% of the total cellular RNA. In addition, there is another type of "invisible interferer" in whole blood samples—Globin mRNA (globin messenger RNA), whose proportion can reach 30%-80% (fluctuating with sample conditions). The combined effect of these two poses multiple obstacles to RNA-seq experiments, making key research areas such as disease biomarker discovery and transcriptome diversity studies extremely challenging.
1. The Double "Obstacles" in RNA-seq Experiments
Both rRNA and Globin mRNA, due to their high abundance, directly impact the analysis results in the following ways:
01. They occupy sequencing resources, leading to insufficient sequencing coverage of mRNA (which already has a very low proportion in total RNA). In particular, low-abundance functional genes may be completely "drowned out";
02. To capture valid information, sequencing depth must be significantly increased. This not only drives up experimental costs but also generates a large amount of redundant data, reducing analysis efficiency;
03. The presence of rRNA interferes with the identification of transcript isoforms (e.g., alternative splicing products), affecting the accurate interpretation of gene expression regulation mechanisms;
04. It limits the analysis of transcriptome diversity and reduces the sequencing space for other transcripts, causing the regulatory functions of low-abundance non-coding RNAs in blood to be overlooked and compromising the comprehensiveness of the research.
Notably, rRNA interference does not need to be addressed in all RNA research: In RNA-cap experiments, since specific probes only focus on capturing the mRNA sequences of target genes, rRNA generally does not interfere with capture efficiency or target sequence analysis, so routine removal is unnecessary. However, in RNA-seq, the effective removal of rRNA and Globin mRNA is a key prerequisite for obtaining reliable and accurate data.
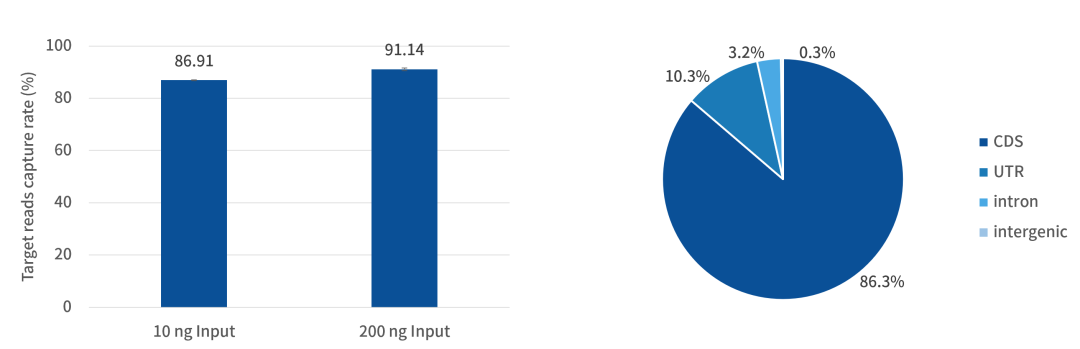
Figure 1. Under the actual capture data of iGeneTech RNA Exome Panel
Even without rRNA and Globin mRNA removal during the library construction step, over 91% of the data can be concentrated in the target regions. Even with a low input amount of 10 ng, more than 86% of the data can still align to the target regions.
2. Addressing Dual Interference: Multiple Technical Approaches to rRNA and Globin mRNA Removal
1. Poly(A) Enrichment Method
This method utilizes the polyA tail of mRNA and employs OligoT probes for hybridization to separate and purify mRNA. It is only applicable to eukaryotic mRNA with a polyA tail and has extremely high requirements for RNA integrity—if the polyA tail of mRNA is broken due to degradation, the mRNA cannot be effectively enriched. More critically, it cannot enrich non-coding RNAs (such as lncRNA and circRNA) that do not have a polyA tail.
2. Probe Hybridization + RNase H Digestion Method
Its principle is to allow specific DNA probes to hybridize with rRNA, then use RNase H to specifically digest the RNA single strand in the DNA-RNA hybrid strands, thereby enriching target RNAs such as mRNA and lncRNA. However, this method involves cumbersome operation steps, requiring additional processes like probe hybridization and enzyme digestion, which prolongs the experimental cycle. Additionally, digestion efficiency is easily affected by factors such as probe specificity and enzyme activity, making it difficult to ensure the stability of removal efficiency. It may also cause certain damage to target RNAs.
3. RiboZero Method
This method uses DNA or RNA probes for hybridization to capture rRNA, which is then removed by adsorption with streptavidin magnetic beads. Although it can remove rRNA to a certain extent, the magnetic bead adsorption process requires multiple washing and centrifugation steps—this complex operation easily leads to RNA loss. Especially in low-input samples (e.g., trace clinical samples), it may affect the efficiency of subsequent library construction. Meanwhile, it cannot achieve targeted removal of Globin mRNA, so it still needs to be combined with other solutions in whole blood sample research, increasing experimental complexity.
4. Reverse Transcription Blocking Probe Method
This method uses reverse transcription blocking probes with high binding affinity to prioritize blocking rRNA and Globin mRNA, preventing reverse transcriptase from accessing the target rRNA and Globin mRNA sites locked by the blocking probes. In this way, it achieves the goal of removing the target RNAs during the reverse transcription process.
3. iGeneTech Dual-Effect Blocking Reagents: 7-Minute Breakthrough to Unlock a New RNA-seq Experience
iGeneTech has developed high-efficiency blocking reagents—IGT Human rRNA Pre-blocking Oligo and IGT Human Globin Pre-blocking Oligo—targeting the Globin and rRNA sequences of human samples, providing a "fast, convenient, and efficient" solution for RNA-seq experiments.
Whether it is disease biomarker discovery in whole blood samples or transcriptome diversity research in FFPE samples, iGeneTech's rRNA and Globin mRNA Blocking Reagents can, with the advantages of "fast, efficient, and flexible", help you break free from the constraints of interfering RNAs, obtain more accurate and valuable RNA-seq data, and facilitate scientific research breakthroughs!
Actual Measurement Data
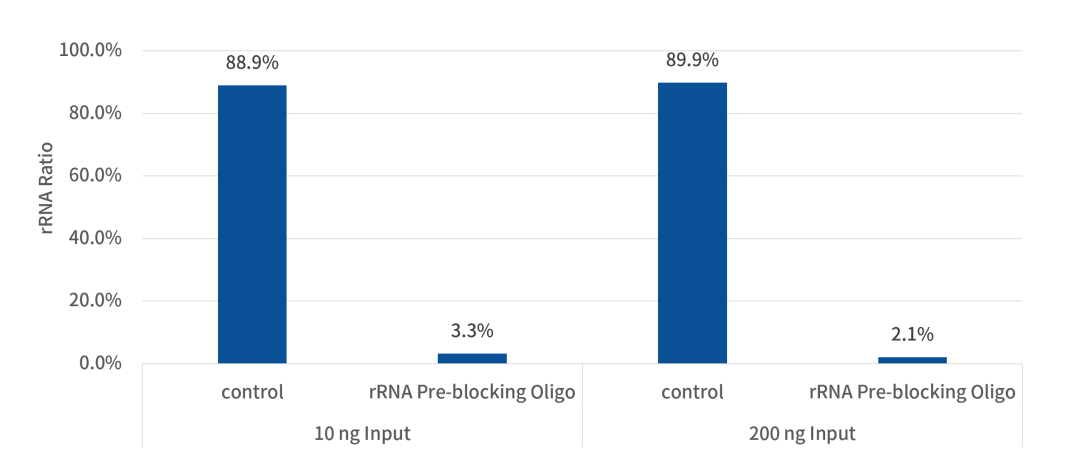
1. rRNA Removal Rate Exceeds 98%
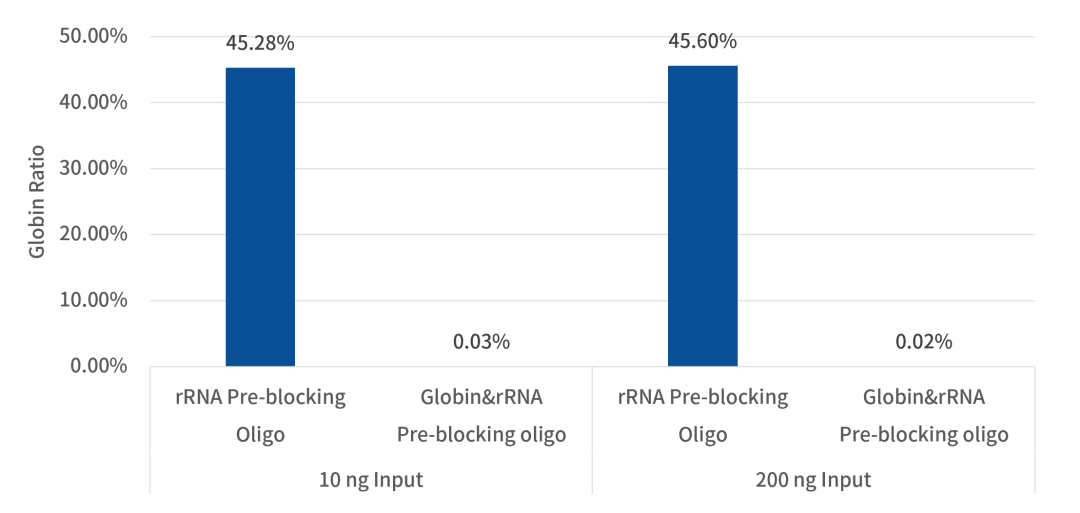
2. Globin mRNA Proportion Reduced to 0.02%
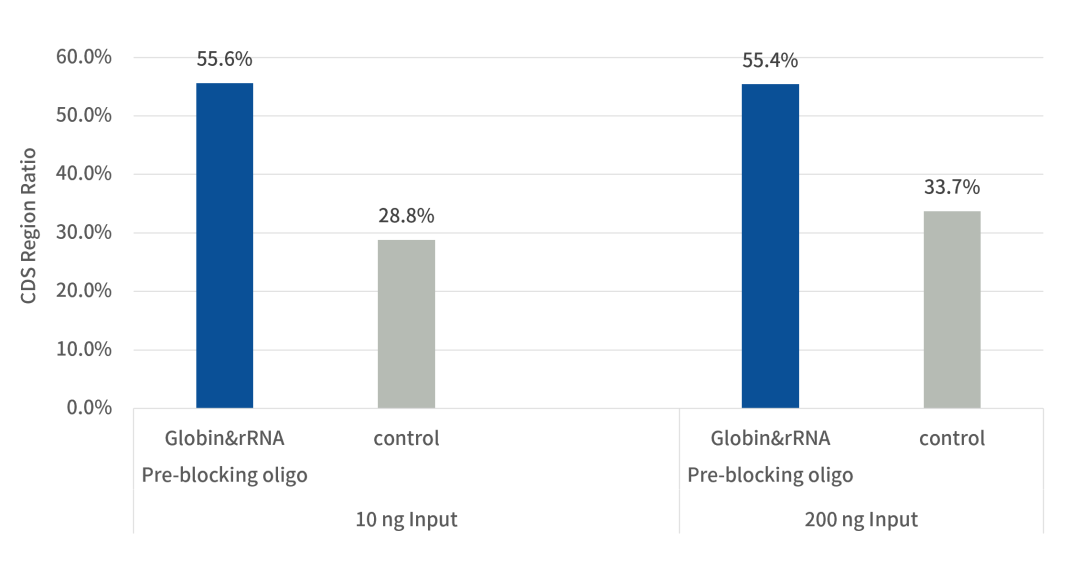
3. The Proportion of CDS Regions Increased by 83%
Product Info

 CN
CN





