Aging is the cumulative decline in systemic physiological function over time, and it serves as the core biological basis driving the occurrence and progression of various age-related chronic diseases. This process varies among individuals, with its dynamic rate synergistically regulated by multiple levels of factors such as genetic background, environmental exposure, lifestyle, and random molecular damage.
Therefore, accurately quantifying the degree of deviation of biological age from chronological age holds crucial translational medical value for identifying individuals at high risk of premature aging, predicting disease susceptibility and mortality risk, and formulating personalized intervention strategies.
DNA methylation is an important epigenetic mechanism through which the body regulates gene expression by controlling the methylation status of CpG sites. The methylation clock, also known as the epigenetic aging clock, is a predictive model for biological age constructed using bioinformatics methods based on DNA methylation data.
This model screens a set of sites highly associated with aging from a large number of CpG sites, predicts an individual’s actual age or aging-related physiological processes by quantifying their methylation status, and the output estimated biological age is referred to as DNA methylation age or epigenetic age¹.
In 2024, an article published by Crimmins in Epigenomics² summarized the four-generation development of methylation clocks:
First-generation Time Clocks
Trained solely based on chronological age, they boast high prediction accuracy but struggle to distinguish health differences among individuals of the same age.
Second-generation Biological Clocks
Integrating mortality and health data, a typical representative is PhenoAge³. It incorporates age and 9 biomarkers including lymphocyte ratio, white blood cell count, and C-reactive protein, enabling prediction of risks for diabetes, cancer, Alzheimer's disease, etc.
Third-generation Clocks
Breaking through the limitation of static assessment, clocks such as DunedinPACE⁴ can dynamically monitor aging rate like a "speedometer." Modeled on 19 biomarkers including blood pressure, lung capacity, high-density lipoprotein, and triglycerides, they predict aging speed.
Fourth-generation Clocks
Combining Mendelian randomization methods, they attempt to distinguish whether methylation changes are the "cause" or "effect" of aging. Focusing on CpG sites that directly drive aging, these clocks aim to reveal the fundamental mechanisms of aging and provide a basis for targeted interventions⁵.
iGeneTech has launched two capture Panels for methylation clock research, empowering precise aging assessment and population-specific model construction:
1. CpG Galaxy Panel
Capture range: 100 Mb genomic region
Clock coverage: Key sites of methylation clock biomarkers such as PhenoAge, Horvath, EpiTOC, and Hannum
2. OmniTrait Methylation Panel
Capture range: 30 Mb genomic region
Clock coverage: Key sites of methylation clock biomarkers such as Horvath, Hannum, Levine, and Bohlin
Synergistic Advantages of the Dual-platform
Both Panels offer extensive CpG site coverage, supporting researchers in independently constructing Chinese population-specific methylation clock models. They meet multi-level needs from exploratory research to clinical translation.
Core Performance Highlights
l Low sample input requirement: A minimum of 5 ng DNA suffices for library construction and capture, making it particularly suitable for precious clinical samples or low-input DNA scenarios.
l Flexible detection: Unlike traditional solid-phase chips that require batch sampling (≥8 samples), this system supports single-sample on-demand sequencing. It achieves rapid "sample-to-data" response and significantly shortens project cycles.
l Cost-effectiveness: Breaking the limitation of single-library single-hybridization, it allows multiplex library pooling for capture. Flexible sample throughput configuration reduces unit monitoring costs and improves reagent utilization.
Technical Performance Validation Data
CpG Galaxy Panel · Technical Reproducibility
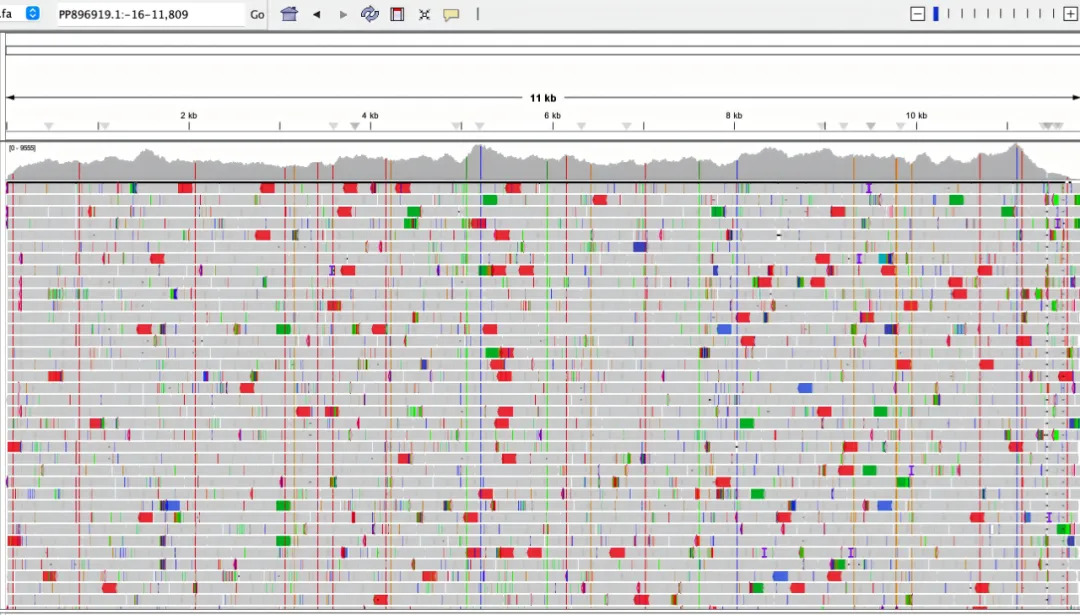
Using 200 ng G304 gDNA, two technical replicates were performed starting from library construction, with a methylation level consistency of R=0.98 between the replicates.
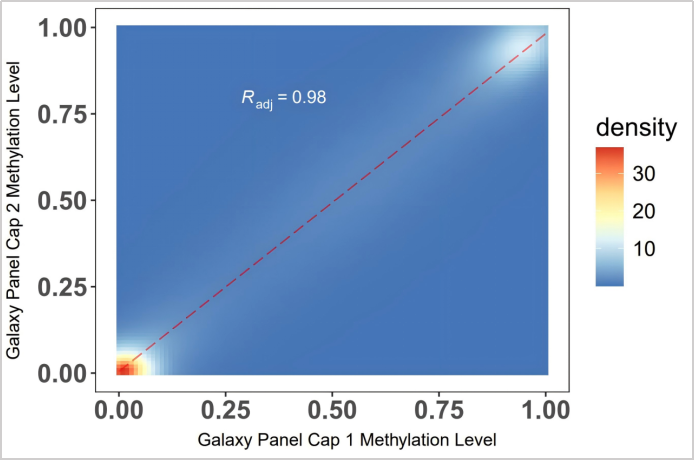
Figure 1. 200 ng G304 gDNA was sonicated, converted using the EpiTect Fast DNA Bisulfite Kit, library-constructed with the methylated double-strand library preparation kit (IGT™ Methyl Fast Library Prep Kit v2.0), captured by the CpG Galaxy Panel, and sequenced on NovaSeq 6000-PE150.
Methylation Level Consistency Between CpG Galaxy Panel and WGBS
200 ng NA12878 gDNA was used for WGBS and CpG Galaxy Panel experiments respectively. Overlapping CpG sites were selected for consistency analysis. The experimental results showed that the consistency between CpG Galaxy Panel and WGBS was R=0.95.
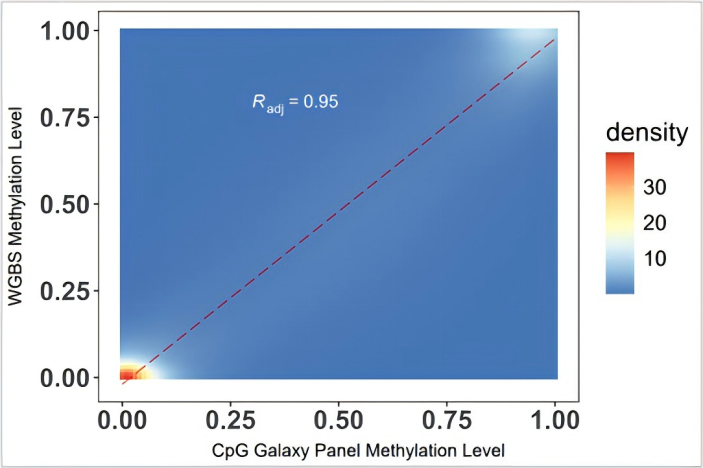
Figure 2. 200 ng NA12878 gDNA was sonicated, converted using the EpiTect Fast DNA Bisulfite Kit, and library-constructed with the methylated double-strand library preparation kit (IGT™ Methyl Fast Library Prep Kit v2.0). WGBS and CpG Galaxy Panel capture were performed respectively, followed by sequencing on NovaSeq 6000-PE150.
OmniTrait Methylation Panel · Technical Reproducibility
Using 50 ng NA12878 DNA, two technical replicates were conducted starting from library construction, with a methylation level consistency of R=0.96 between the replicates.
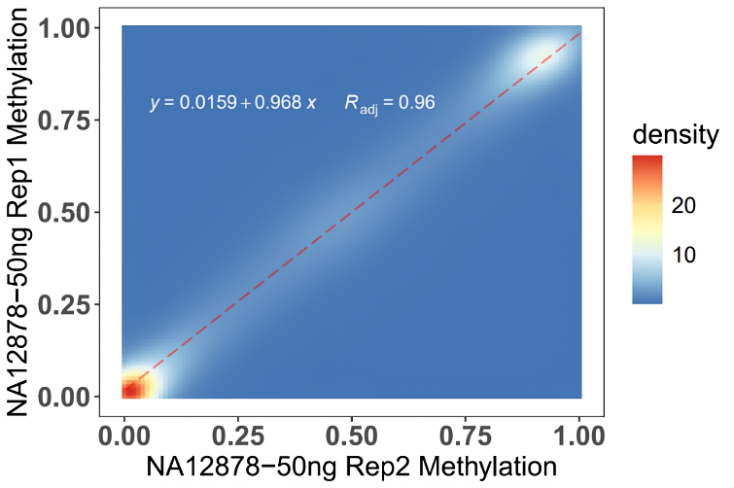
Figure 3. 50 ng NA12878 DNA was sonicated, converted using the EpiTect Fast DNA Bisulfite Kit, library-constructed with the methylated double-strand library preparation kit (IGT Methyl Fast Library Prep Kit v2.0), captured by the OmniTrait Methylation Panel, and sequenced on NovaSeq 6000-PE150.
OmniTrait Methylation Panel vs. WGBS · Methylation Level Consistency
200 ng NA12878 gDNA was used for WGBS and OmniTrait Methylation Panel experiments respectively. Overlapping CpG sites were selected for consistency analysis. The experimental results showed that the consistency between OmniTrait Methylation Panel and WGBS was R=0.95.
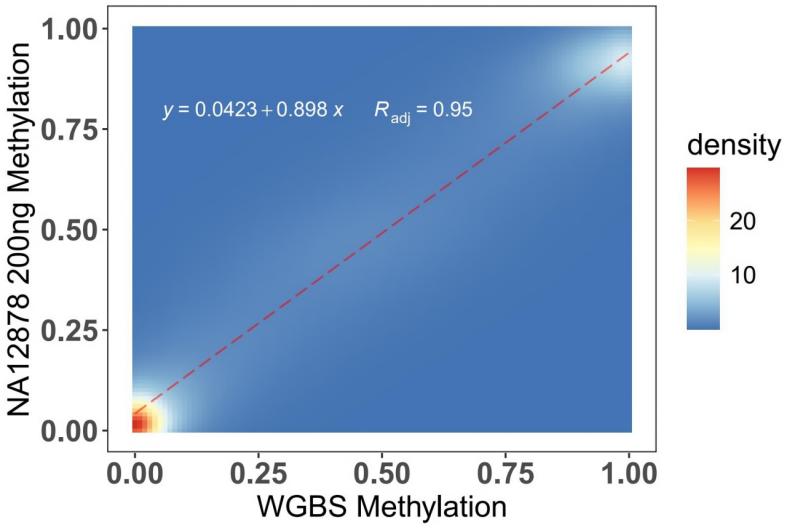
Figure 4. 200 ng of NA12878 gDNA was sonicated, converted using the EpiTect Fast DNA Bisulfite Kit, and library-constructed with the methylated double-strand library preparation kit (IGT™ Methyl Fast Library Prep Kit v2.0). It was then subjected to WGBS and OmniTrait Methylation Panel capture separately, followed by NovaSeq 6000-PE150 sequencing.
References
1. Zhu ZW, Cheng SS, Cheng X, et al. Research and application of aging clocks based on multi-omics data[J]. Chinese Journal of Epidemiology, 2024, 45(9): 1291-1301. DOI: 10.3760/cma.j.cn112338-20240424-00215
2. Crimmins EM, Klopack ET, Kim JK. Generations of epigenetic clocks and their links to socioeconomic status in the Health and Retirement Study. Epigenomics. 2024;16(14):1031-1042. doi: 10.1080/17501911.2024.2373682. Epub 2024 Jul 18. PMID: 39023350; PMCID: PMC11404624.
3. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L, Horvath S. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018 Apr 18;10(4):573-591. doi: 10.18632/aging.101414. PMID: 29676998; PMCID: PMC5940111.
4. Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E, Harrington HL, Houts R, Kothari M, Kwon D, Mill J, Schwartz J, Vokonas P, Wang C, Williams BS, Moffitt TE. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022 Jan 14;11:e73420. doi: 10.7554/eLife.73420. PMID: 35029144; PMCID: PMC8853656.
5. Ryan CP, Belsky DW. Epigenetic clock work ticks forward. Nat Aging. 2024 Feb;4(2):170-172. doi: 10.1038/s43587-024-00570-x. PMID: 38291215.
Product Info
Product Name | Speci. | Cat. No |
OmniTrait Methylation Panel OmniTrait Methylation Panel | 4 rxn | PE4000560 |
24 rxn | PE4000565 |
Mouse CGI Panel Mouse CGI Panel | 4 rxn | PE4000280 |
24 rxn | PE4000285 |
CpG Galaxy Panel | 24 rxn | PE4000175 |
CGI Panel | 24 rxn | PB3000235 |
MCED Panel MCED Panel | 4 rxn | PE4000350 |
24 rxn | PE4000355 |
TargetSeq One® BisCap® Hyb & Wash Kit with Eco Universal Blocking Oligo (for Illumina) | 24 rxn | B30334 |
TargetSeq One® BisCap® Hyb & Wash Kit with Eco Universal Blocking Oligo (for MGI DI) | 24 rxn | B30354 |
TargetSeq One® BisCap® Hyb & Wash Kit with Eco Universal Blocking Oligo (for Illumina ssDNA Library) | 24 rxn | B30364 |
IGT® ssDNA Library Prep Kit | 24 rxn | C10914 |
IGT® Methyl Fast Library Prep Kit v2.0 | 24 rxn | B30014 |
IGT® Methyl Adapter & UDI Primer 1 - 96 (for Illumina, plate) | 96 rxn | B30022 |
IGT® Methyl Adapter & UDI Primer 1 - 96 (for MGI, plate) | 96 rxn | B30172 |

 CN
CN