DNA cytosine methylation is currently the most extensively studied epigenetic modification. In mammalian genomes, this modification primarily occurs at CpG sites (except in neuronal cells and embryonic stem cells). However, in plants and some invertebrates, non-CpG (CHG/CHH) methylation is also prevalent.
Leveraging its own liquid-phase capture technology, iGeneTech has developed targeted capture methylation technology platforms (MethCap and BisCap) for specific genomic regions. These platforms seamlessly integrate targeted capture and methylation sequencing. Depending on research needs, the two technologies—MethCap and BisCap—can be flexibly selected to study the methylation status in any target genomic region.
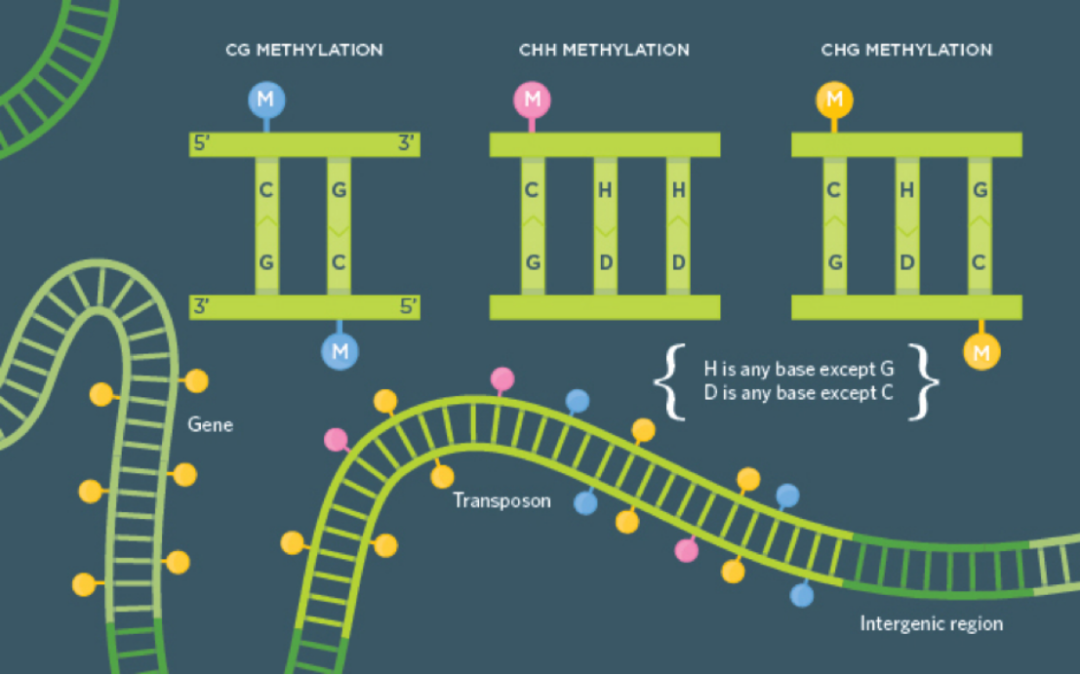
Three Types of DNA Methylation: CG, CHG, CHH (https://www.the-scientist.com/infographic-plant-methylation-basics-32112)
Technical Workflow
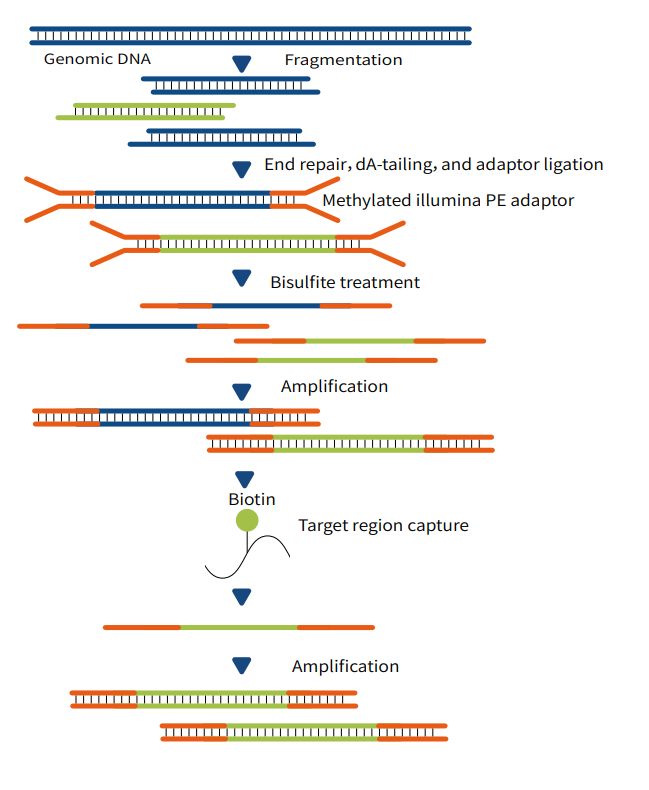
MethCap Technology
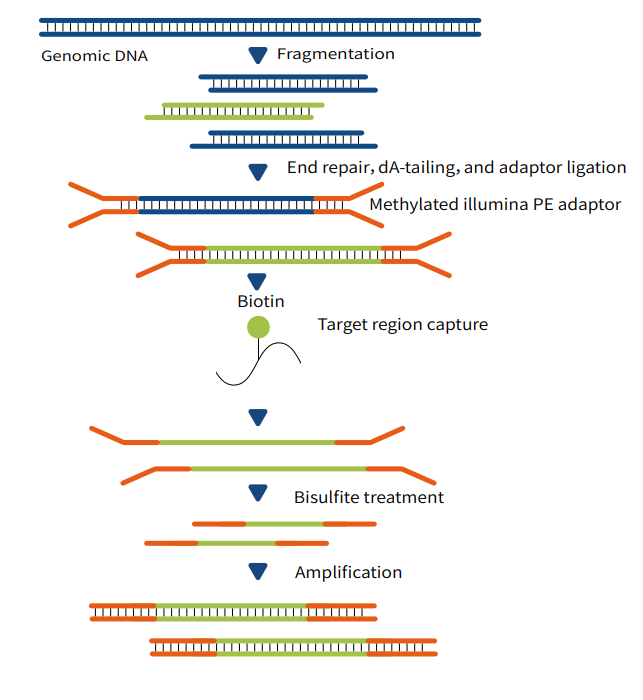
"Capture-First, Conversion-Later": This technology adopts a workflow where target genomic DNA fragments are first captured, followed by bisulfite or enzymatic conversion. The specific steps are as follows:
First, construct a next-generation sequencing (NGS) pre-library;
Second, perform liquid-phase capture to obtain the target genomic regions of interest;
Third, carry out conversion using either bisulfite or enzymatic methods;
Finally, conduct high-throughput sequencing to obtain single-base resolution results for DNA methylation detection.
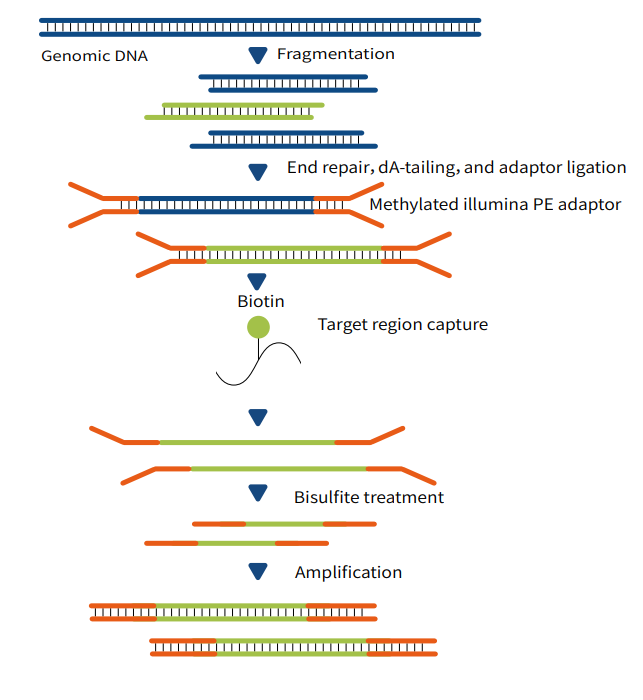
Figure 1 MethCap Technical Workflow
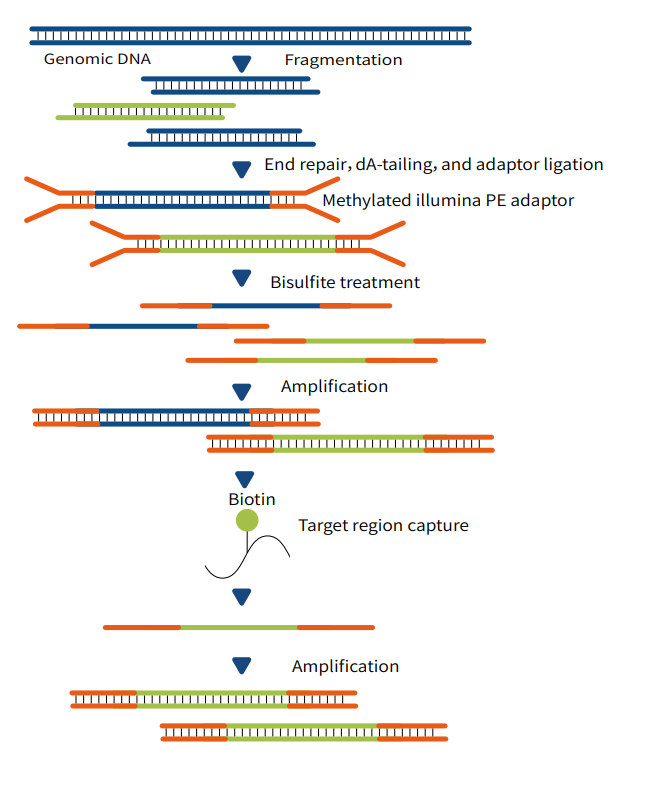
BisCap Technology
"Conversion-First, Capture-Later": This workflow first performs bisulfite or enzymatic conversion on the whole genome, then uses DNA probes to capture the target genomic regions of interest to users, thereby enabling the study and analysis of DNA methylation status within these target regions.
Applicable for DNA with low input amount or high degradation
DNA with high input amount
Selection of Application Scenarios
Basis for Technical Workflow Selection
Through the above two technical workflows: MethCap and BisCap, users can conduct various methylation-related scientific research. So, which technical workflow should users choose for actual projects? We recommend selection based on species, research objectives, and DNA input amount:
The MethCap platform is not limited by species and can detect CpG, CHG, and CHH methylation levels, but requires a DNA library construction input amount of≥1μg.
The BisCap platform is suitable for detecting species with extremely low CHG and CHH methylation levels, such as CpG site methylation in mammals like humans and mice, with low input amount requirements: gDNA≥50ng, cfDNA≥5ng.
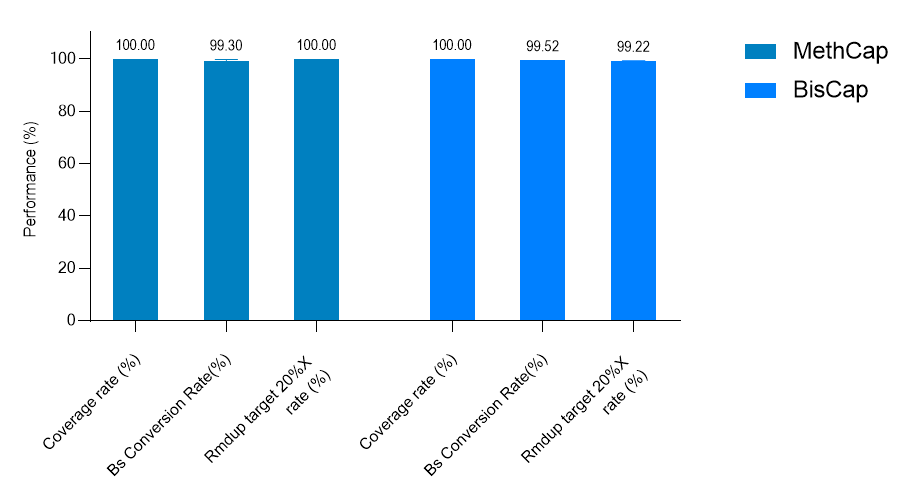
Data Metrics Display for MethCap and BisCap
MethCap was tested using a 3 Kb Panel, while BisCap was tested using a 9 Kb Panel. The resulting data metrics are shown in the figure below: the target region coverage, bisulfite (BS) conversion rate, and uniformity are all excellent. The average post-duplication depths of MethCap and BisCap are 2301× and 3379×, respectively.
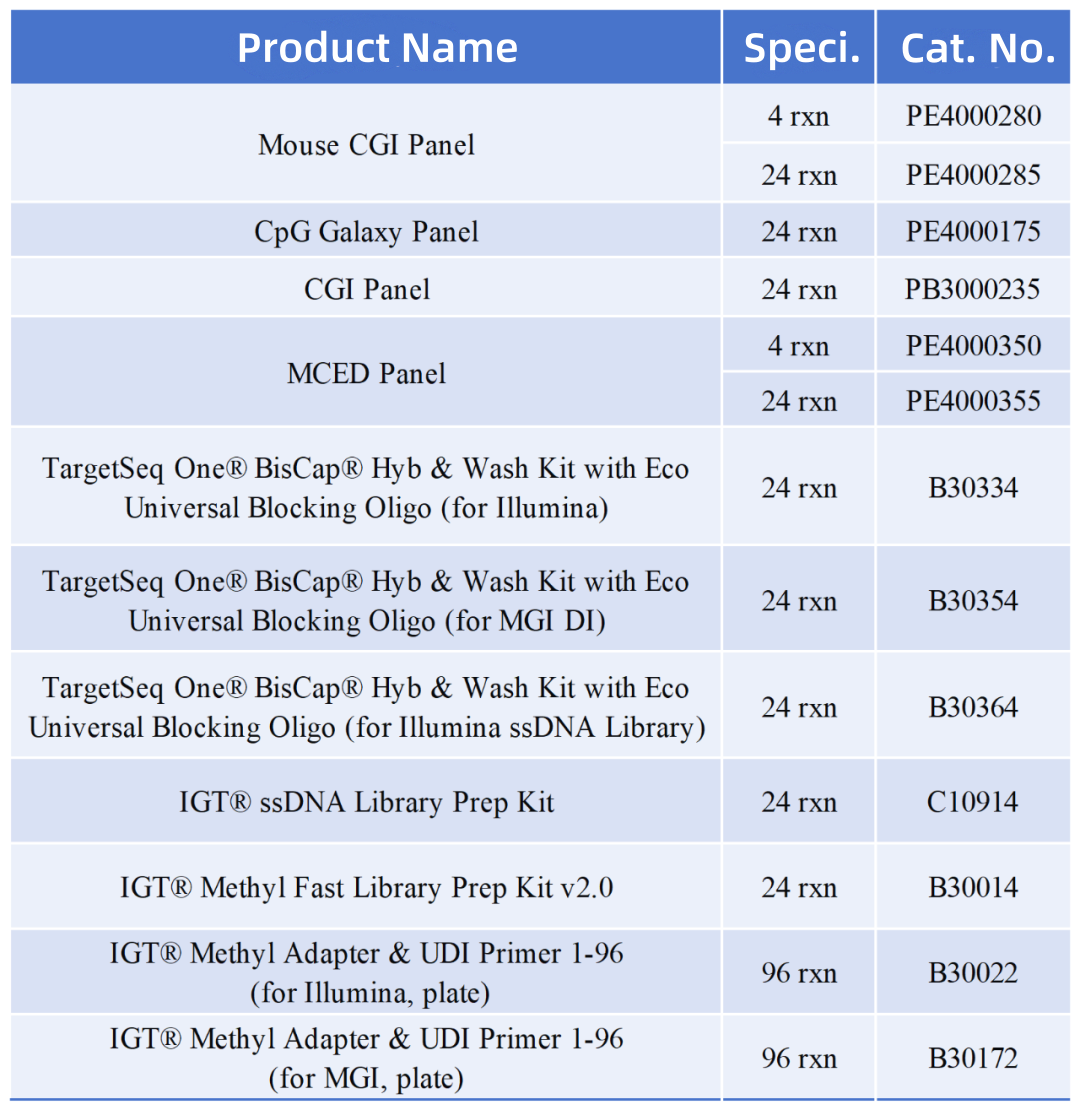
Product Info

 CN
CN