Core Definition
Ultraconserved Elements (UCEs) are specific regions in biological genomes with extremely high sequence conservation, existing even among species that are evolutionarily distant—for example, birds and humans share many UCEs.
UCEs were first reported by Gil Bejerano et al. in 2004 [1]. Subsequent studies identified UCEs in multiple taxonomic groups beyond the initial group used to characterize these genomic elements [2]. Miller et al. further discovered more highly conserved regions through comparative analysis of genomes from 27 vertebrate species [3].
Application Directions
Phylogenetics and Evolution: Construct phylogenetic trees across distantly related species, and decipher deep phylogenetic relationships, rapid radiation, and adaptive evolution mechanisms.
Species Classification and Identification: Assist in automated species classification, resolve taxonomic disputes among morphologically similar species, and standardize classification criteria.
Population Genetics: Analyze population structure, genetic diversity, and differentiation processes, and detect population hybridization and gene flow dynamics.
Genomics and Functional Research: Facilitate genome assembly anchoring of non-model species, and explore functional mechanisms related to non-coding RNAs.
Technical Workflow
1. UCE Locus Identification
First, conduct comparative analysis of multi-species genomic sequences to systematically scan and screen highly conserved regions with 95%-100% sequence identity. Subsequently, combine user-defined filtering criteria such as length thresholds to achieve accurate delineation of UCE loci.
2. Probe Design and Library Preparation
Design specific liquid-phase capture probes targeting the set of UCEs shared by the target taxa. Simultaneously construct DNA libraries, with the recommended library insert size controlled between 300~600 bp.
3. Target Enrichment and Sequencing
Adopt liquid-phase capture technology for UCE-targeted enrichment and high-throughput sequencing of DNA libraries. Sequencing strategies such as PE150, PE250, or PE300 can be selected based on research needs to ensure the coverage and integrity of sequence data.
4. Data Analysis Pipeline
After quality control and filtering of sequencing data, use professional software tools such as phyluce to sequentially complete analytical steps including sequence assembly, UCE locus mapping, and sequence alignment, thereby supporting downstream research on scientific questions such as phylogenetic reconstruction.
Application Case
Case: Combined Application of UCE Capture and Machine Learning
Algorithms in Nematode Phylogenetic Research
Nematodes are one of the most diverse animal taxa, with an estimated 1 million species, but only approximately 28,000 have been described morphologically.
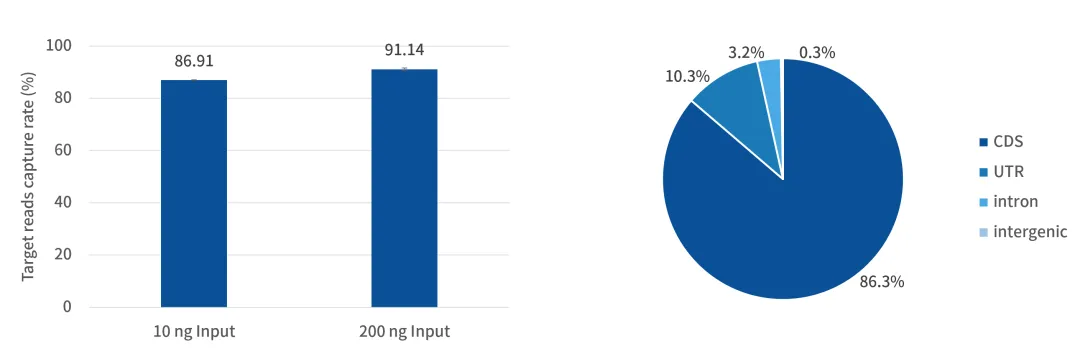
Addressing the limitations of nematode morphological classification and traditional molecular markers, researchers designed a dedicated set of UCE probes for Panagrolaimidae and Rhabditidae for the first time. Combined with target capture technology (library fragment size: 310~350 bp, iGenetech NovaSeq 6000 PE151 sequencing) and existing genomic data, they systematically verified the universality of UCEs in these two nematode families and their potential for phylogenetic analysis.
Meanwhile, machine learning technology was used to achieve automatic sample classification and feature selection. The results showed that high-precision genus-level classification can be completed by identifying only a small number of high-information loci, providing an efficient and reliable new solution for nematode taxonomy research [4].
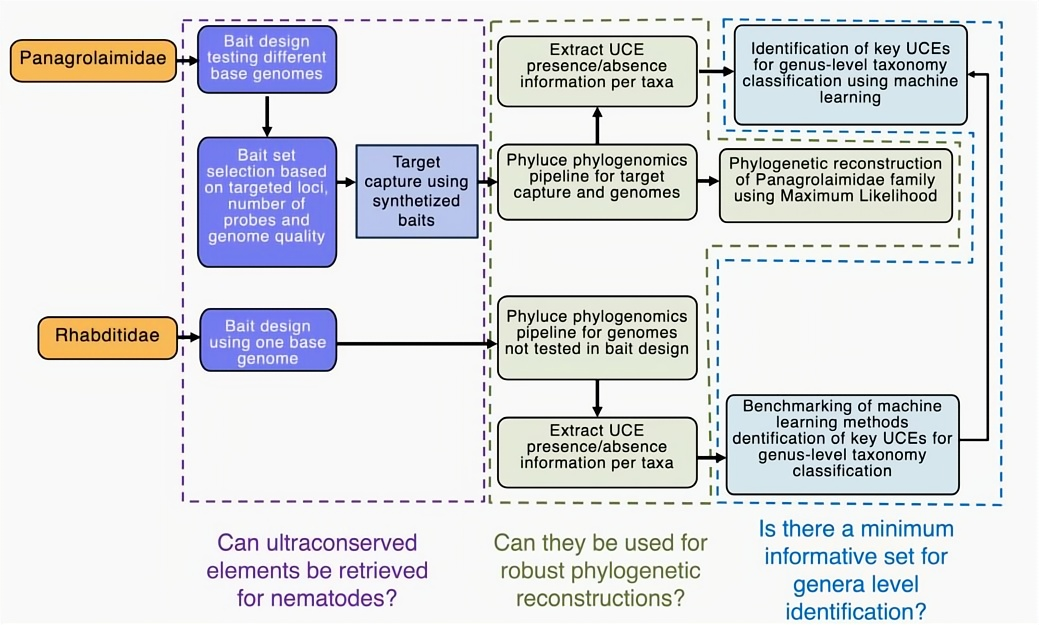
Figure 1. Overview of the workflow implemented for nematode UCE research [4].
Product Information
www.ultraconserved.org is a professional academic resource platform for UCEs research. It is committed to openly sharing UCE-related sequence information and supporting research tools with researchers worldwide, facilitating research in the fields of phylogenomics, phylogeography, and population genetics.
Product Name | Speci. | Cat. No |
Arachnida UCE 1.1K v1 | 24 rxn | PH2016235 |
96 rxn | PH2016232 |
Coleoptera UCE 1.1K v1 | 24 rxn | PH2014785 |
96 rxn | PH2014782 |
Diptera UCE 2.7K v1 | 24 rxn | PH2016225 |
96 rxn | PH2016222 |
Hemiptera UCE 2.7K v1 | 24 rxn | PH2016175 |
96 rxn | PH2016172 |
Hymenoptera 2.5K v2 | 24 rxn | PH2016205 |
96 rxn | PH2016202 |
Anthozoan UCE 1.7K v1 | 24 rxn | PH2016215 |
96 rxn | PH2016212 |
Actinopterygians UCE 0.5K v1 | 24 rxn | PH2006675 |
96 rxn | PH2006672 |
Acanthomorphs UCE 1K v1 | 24 rxn | PH2016185 |
96 rxn | PH2016182 |
Tetrapods UCE 2.5K v1 | 24 rxn | PH2016195 |
96 rxn | PH2016192 |
Tetrapods UCE 5K v1 | 24 rxn | PH2010095 |
96 rxn | PH2010092 |
References
[1] Bejerano G , Pheasant M , et al.Ultraconserved Elements in the Human Genome[J].Science, 2004, 304(5675):1321-1325.DOI:10.1126/science.1098119.
[2] Siepel,A.Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes[J].Genome Research, 2005, 15(8):1034-1050.DOI:10.1101/gr.3715005.
[3] Miller, Webb et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser.Genome research vol. 17,12 (2007): 1797-808. doi:10.1101/gr.6761107.
[4] Villegas, Laura , et al. Ultraconserved Elements and Machine Learning Classifiers Enable Robust Phylogenetics and Taxonomy in Model and Non‐Model Nematodes. Molecular Ecology Resources 25.8(2025).

 CN
CN