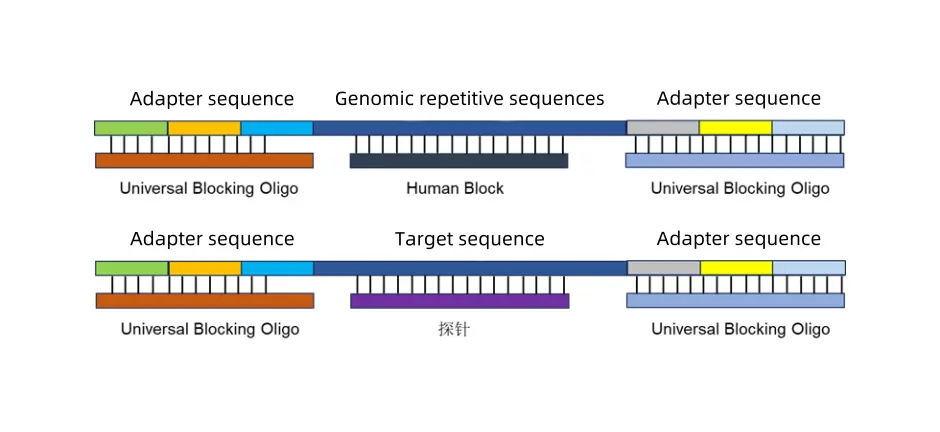
In the work of infectious disease prevention and control as well as emergency monitoring, the accurate identification and genotyping analysis of pathogens are key to achieving scientific prevention and control.
With ten years of accumulation in capture technology, iGeneTech has launched whole-genome capture and sequencing solutions for CDCs (Centers for Disease Control and Prevention) and public health laboratories, providing strong technical support for pathogen monitoring, outbreak tracing, and drug resistance analysis.
01 Intestinal Pathogens: Supports Outbreak Tracing and Virulence Detection
It covers key monitoring targets of intestinal infectious diseases, including Vibrio cholerae, Salmonella, Shigella, diarrheagenic E. coli, Campylobacter, and Yersinia. The solution enables:
√ Simultaneous detection of serotypes and virulence genes
√ Rapid tracing analysis of outbreak events
√ Combined detection of multiple samples
Applicable to foodborne disease monitoring, intestinal epidemic tracing, and local epidemic trend assessment.
02 Respiratory Pathogens: Improves Epidemic Monitoring and Genotyping Efficiency
For respiratory pathogens such as Neisseria meningitidis, Haemophilus influenzae, Streptococcus pneumoniae, Legionella, and Bordetella pertussis, iGeneTech provides a mature whole-genome capture probe solution, supporting:
√ High coverage of whole-genome sequencing (>99% of regions)
√ Multi-dimensional feature identification: serotypes, drug resistance genes, and virulence factors
√ Batch application and standardized output in regional sentinel laboratories
Applicable to the monitoring of epidemic strains of meningococci, vaccine efficacy evaluation, and epidemiological research on respiratory pathogens.
03 Special and Highly Pathogenic Pathogens: Builds a High-Security Prevention Barrier
For pathogens such as Bacillus anthracis, Brucella, Corynebacterium diphtheriae, Listeria monocytogenes, Klebsiella pneumoniae, Mycoplasma pneumoniae, Chlamydia pneumoniae, and Chlamydia psittaci, the iGeneTech solution provides:
√ Integrated analysis of whole-genome capture and virulence feature identification
√ Comprehensive analysis of drug resistance genes, virulence factors, and evolutionary lineages
√ Support for tracing and comparison of epidemic strains across regions
Meets the needs of highly pathogenic pathogen detection, emergency analysis of public health emergencies, and biosafety monitoring.
Technical Advantages: Empowering the CDC System
iGeneTech’s whole-genome capture system for pathogenic microorganisms has been verified and applied in CDC laboratories in many regions, with the following features:
√ High Sensitivity — Maintains high coverage even for low-copy samples
√ High Parallelism — Enables simultaneous enrichment of multiple pathogens
√ High Compatibility — Compatible with multiple sequencing platforms such as Illumina and MGI
√ High Standards — Complies with CDC data standards for tracing analysis
Scientific prevention and control start with precise capture.
Product Info


 CN
CN