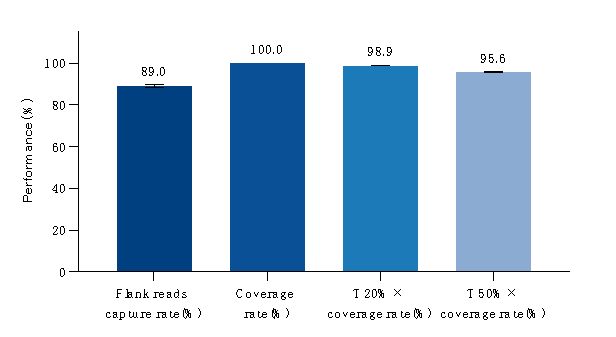
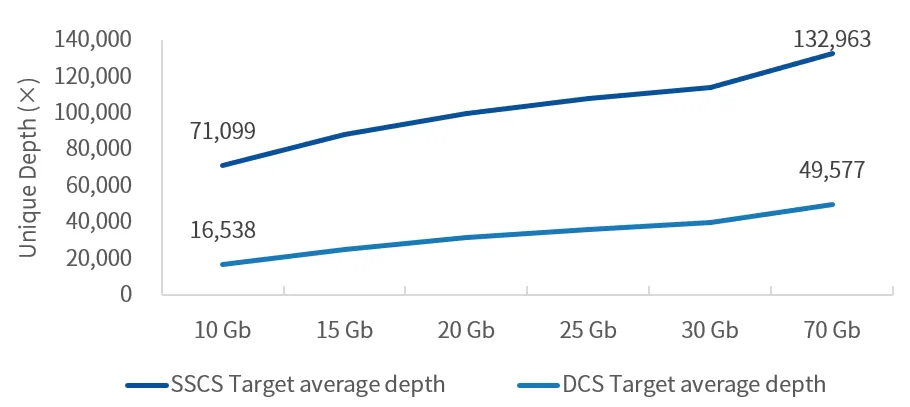
Overview
Based on TargetSeq® hybrid capture sequencing technology, it covers 641 genes related to targeted therapy for solid tumors and tumor genetic susceptibility. Additionally, it includes intronic regions of 38 gene hot spot fusions, 15 classic microsatellite loci, and 219 chemotherapy-related loci. It enables one-time detection of variant types such as point mutations, fusions, copy number variations, and insertions/deletions, as well as TMB and MSI analyses, providing references for tumor targeted therapy, immunotherapy, and risk prediction of hereditary tumors, among others.

 CN
CN