Overview
TargetSeq® Solid Tumor Mid Panel covers 122 gene coding regions related to solid tumors, 10 hotspot fusion gene intron regions, 19 classic microsatellite sites, and 219 chemotherapy-related sites.
TargetSeq® Solid Tumor Mid Panel covers 122 gene coding regions related to solid tumors, 10 hotspot fusion gene intron regions, 19 classic microsatellite sites, and 219 chemotherapy-related sites.
Panel Number | T1288V1 |
Technical Platform | TargetSeq® Hybridization Capture Sequencing |
Coverage Size | 438.3 kb |
Reference Database | RefSeq |
Reference Genome | GRCh37 (hg19) |
Coverage | CDS region of 122 genes, 10 hotspot fusion gene intron regions, 19 MSI and 219 chemotherapy-related sites. |
Storage | -20℃±5℃ |
Sequence Platform | Illumina / MGI |
Recommend Sequencing Read Length | PE150 |
Recommend Sequencing Data Size and Depth | 1.5Gb/1,000X |
Selected coverage of 122 key pan-cancer genes;
Simultaneous detection of various variant types including SNP/Indel, CNV, Fusion, and MSI.
Coupled with high-efficiency library preparation and hybrid capture reagents, it accurately targets low-abundance variants.
Capture Performance Test
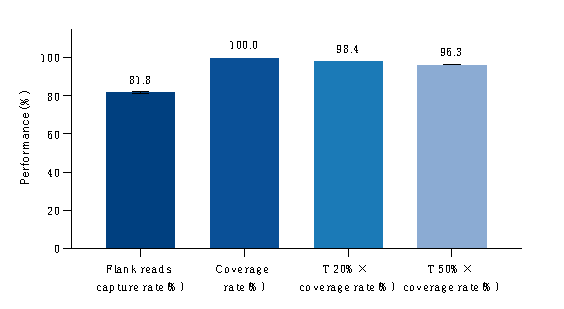
Figure 1. Test of Capture Data Performance of gDNA Reference Standards. gDNA positive reference standards (Shuimu Jiheng, PSC500) were used to construct libraries with an input amount of 100 ng, followed by hybrid capture and sequencing on the Illumina NovaSeq 6000 with PE150.
Table 1. Detection results of mutation sites in gDNA positive reference standards
Gene | Variant | Expected Allelic Frequency | Reported Allelic Frequency |
EGFR | L858R | 1.0% | 0.9% |
KRAS | A146T | 1.0% | 1.4% |
NRAS | Q61K | 1.0% | 0.9% |
EGFR | T790M | 2.0% | 1.9% |
EGFR | E746_A750del | 2.0% | 1.5% |
FLT3 | I836del | 2.0% | 1.8% |
KIT | D816V | 2.0% | 2.3% |
KRAS | G12D | 2.0% | 1.6% |
EGFR | A767_V769dup | 3.0% | 1.9% |
EGFR | G719S | 4.0% | 4.5% |
EML4-ALK | Fusion | 5.0% | 3.6% |
| Product Name | Panel Number | Set | Cat. No |
TargetSeq® Solid Tumor Mid Panel | T1288V1 | 16 rxn | PH2002101 |
| 96 rxn | PH2002102 |