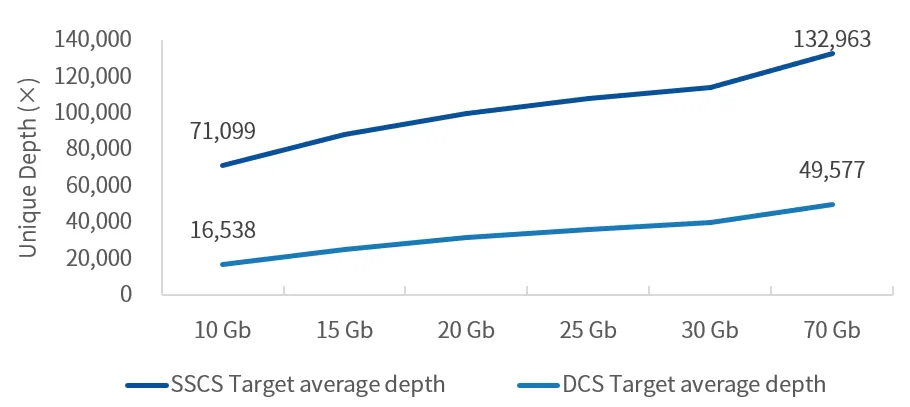
Capture performance testing
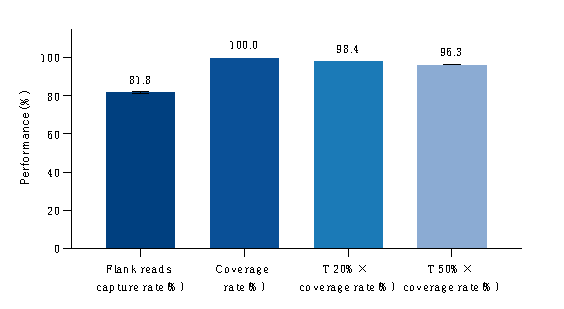
Figure 1. Capture performance test of gDNA reference standards.
Note: Tested with gDNA reference standards. Libraries were constructed using the IGT® Fast Library Prep Kit v2.0, coupled with the TargetSeq One® Hyb & Wash Kit v3.0 hybridization and washing system, and sequenced on the Illumina NovaSeq 6000 with PE150 (paired-end 150).
Mutation Detection
Table 1. Detection results of mutation sites in gDNA positive reference standards
| Gene
| Variant | Expected allelic frequency | Reported allelic frequency |
EGFR | L858R | 1.0% | 1.7% |
KRAS | A146T | 1.0% | 0.7% |
NRAS | Q61K | 1.0% | 0.9% |
EGFR | T790M | 2.0% | 2.3% |
EGFR | ΔE746_A750 | 2.0% | 1.1% |
KRAS | G12D | 2.0% | 1.3% |
EGFR | V769_D770insASV | 3.0% | 1.4% |
EGFR | G719S | 4.0% | 4.3% |
KRAS | G13D | 4.0% | 3.3% |
EML4-ALK | Fusion | 5.0% | 2.4% |
CD74-ROS1 | Fusion | 6.0% | 2.9% |
BRAF | V600E | 7.0% | 6.3% |
PIK3CA | H1047R | 7.0% | 4.7% |
Note: Comparative analysis results of mutation site detection in positive reference standards. Libraries were constructed using gDNA reference standards (Jingliang, cat. GW-OPSM003) with an input amount of 50 ng, sequenced on the Illumina NovaSeq 6000 with PE150. When the depth after deduplication was ~1,000× (1 Gb raw bases), all positive mutation sites were detected, which is consistent with the expected results.
Table 2. Detection results of mutation sites in ctDNA positive reference standards
Gene | Variant | Expected allelic frequency | Reported allelic frequency |
NRAS | Q61K | 0.5% | 0.6% |
NRAS | A59T | 0.5% | 0.8% |
PIK3CA | E545K | 0.5% | 0.7% |
EGFR | ΔE746 - A750 | 0.5% | 0.6% |
EGFR | V769 - D770insASV | 0.5% | 0.6% |
EGFR | T790M | 0.5% | 0.6% |
EGFR | L858R | 0.5% | 0.6% |
KRAS | G12D | 0.5% | 0.8% |
Note: Comparative analysis results of mutation site detection in positive reference standards. Libraries were constructed with an input amount of 10 ng using cfDNA reference standards (Horizon, cat. HD780), sequenced on the Illumina NovaSeq 6000 with PE150. When the depth after deduplication was ~10,000× (4 Gb raw bases), all positive mutation sites were detected, which is consistent with the expected results.

 CN
CN