Highlight 1: Triple-layer Safeguard of Backbone Probes for Comprehensive CNV Detection
To address diverse CNV detection needs, AIExome® V5-Inherit innovatively constructs a three-tier CNV detection network, improving the CNV resolution for key genes to the single-exon level and supporting genome-wide detection of CNVs ≥1 Mb. This achieves comprehensive CNV variation coverage from fine-scale to macroscopic perspectives.
First Layer of Safeguard: High density backbone for VIP Genes
In accordance with expert consensus and guidelines, and integrating the opinions of multiple clinical testing and interpretation experts, AIExome® V5-Inherit incorporates special designs for multiple genes associated with inherit diseases. This significantly improves the detection rates for hereditary disorders such as Duchenne muscular dystrophy, thalassemia, spinal muscular atrophy, and congenital adrenal hyperplasia.
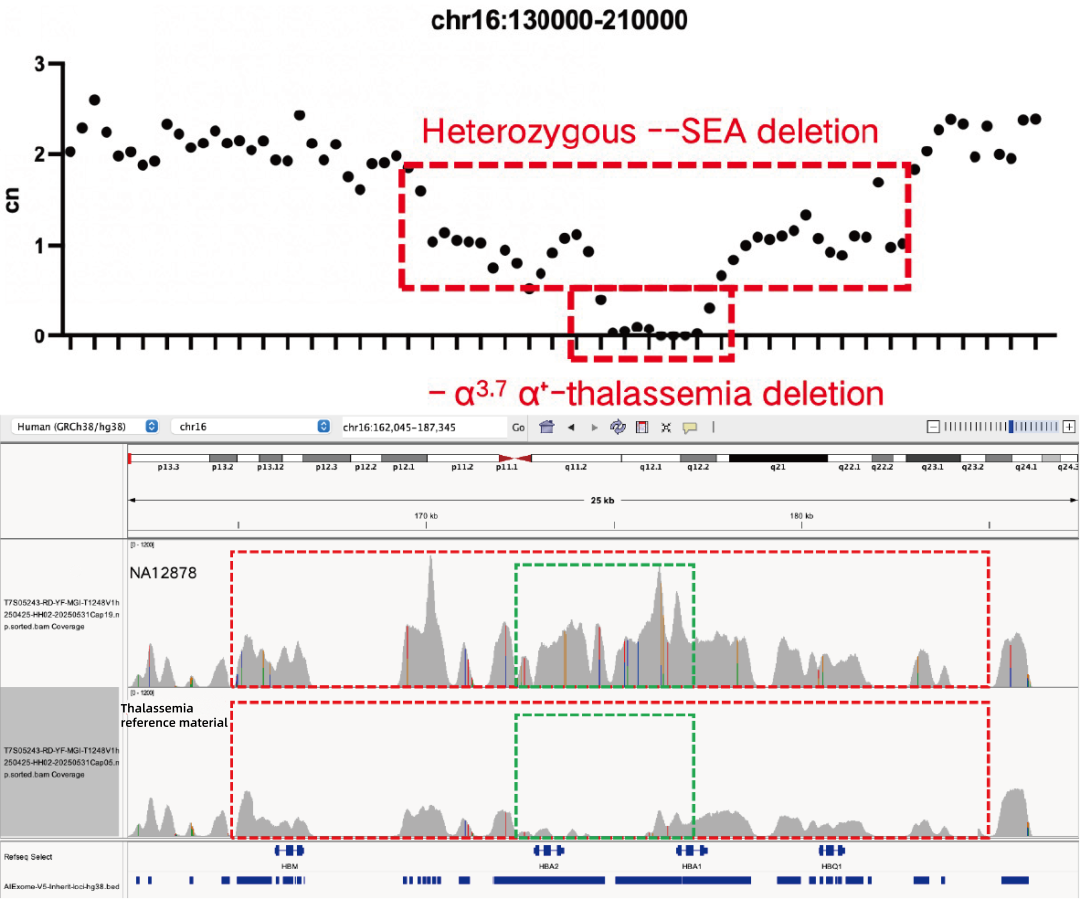
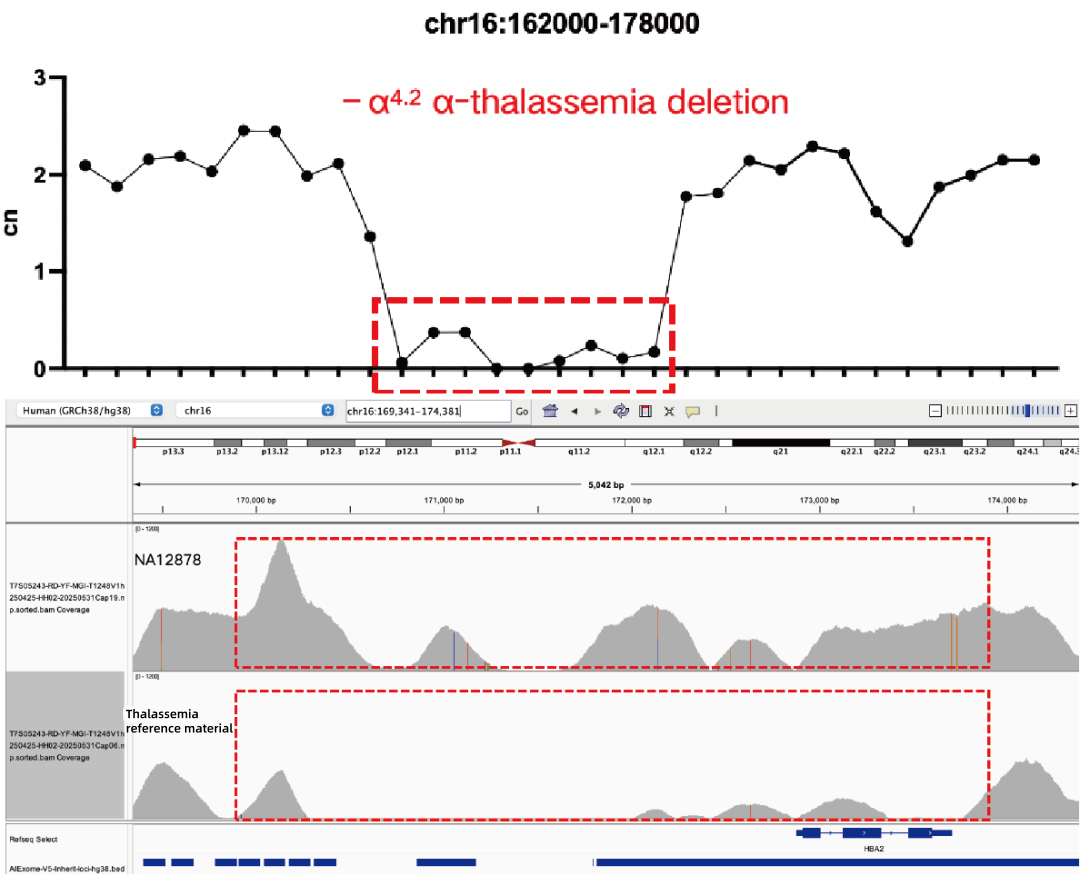
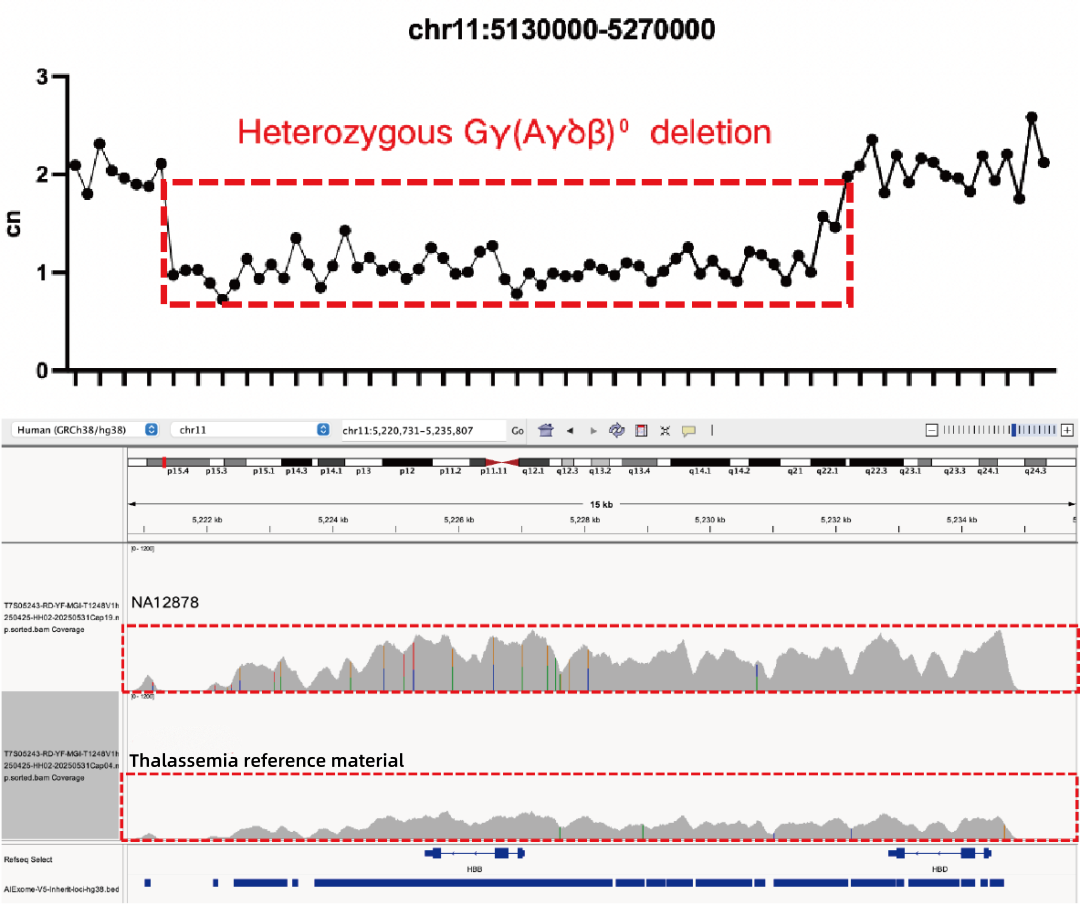
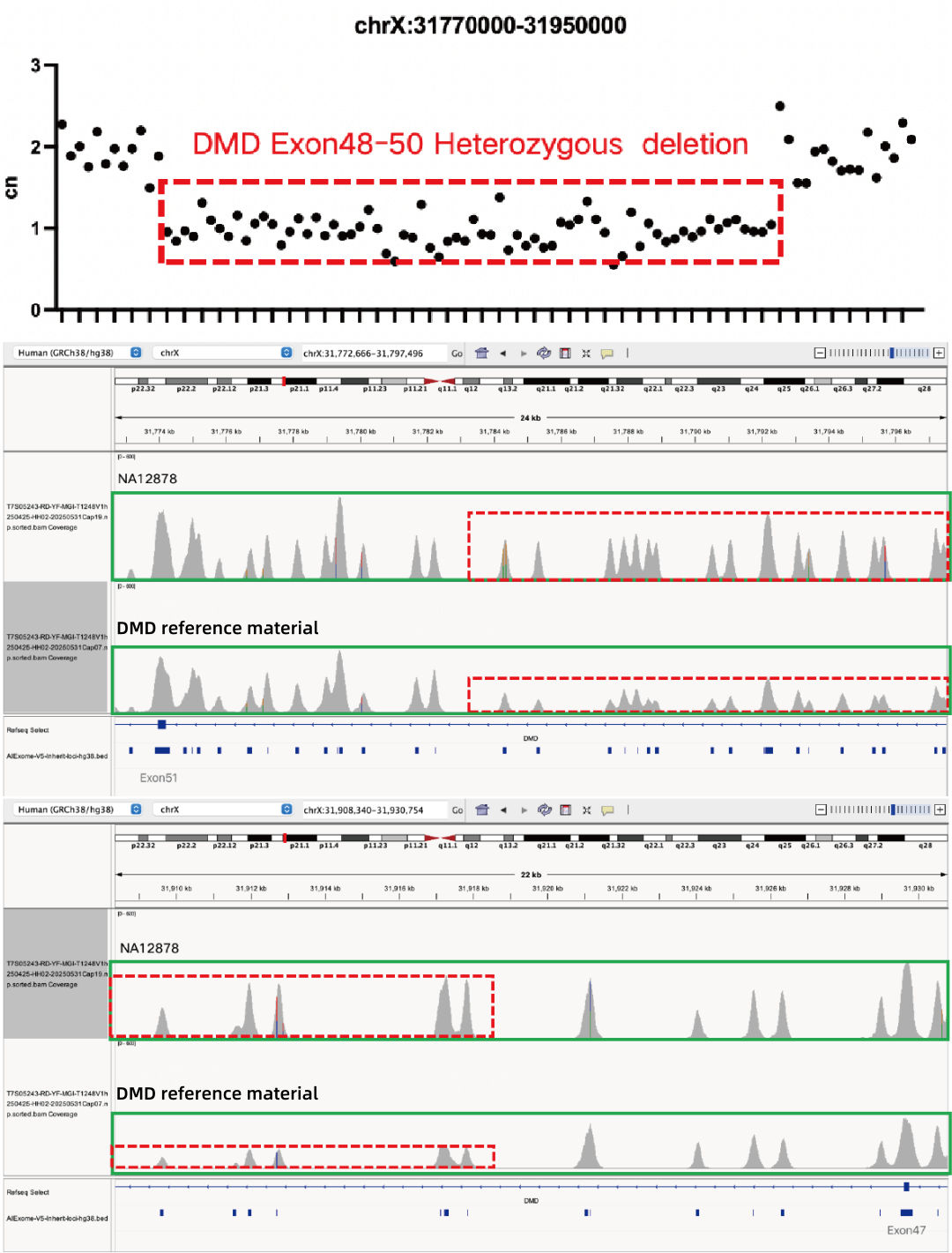
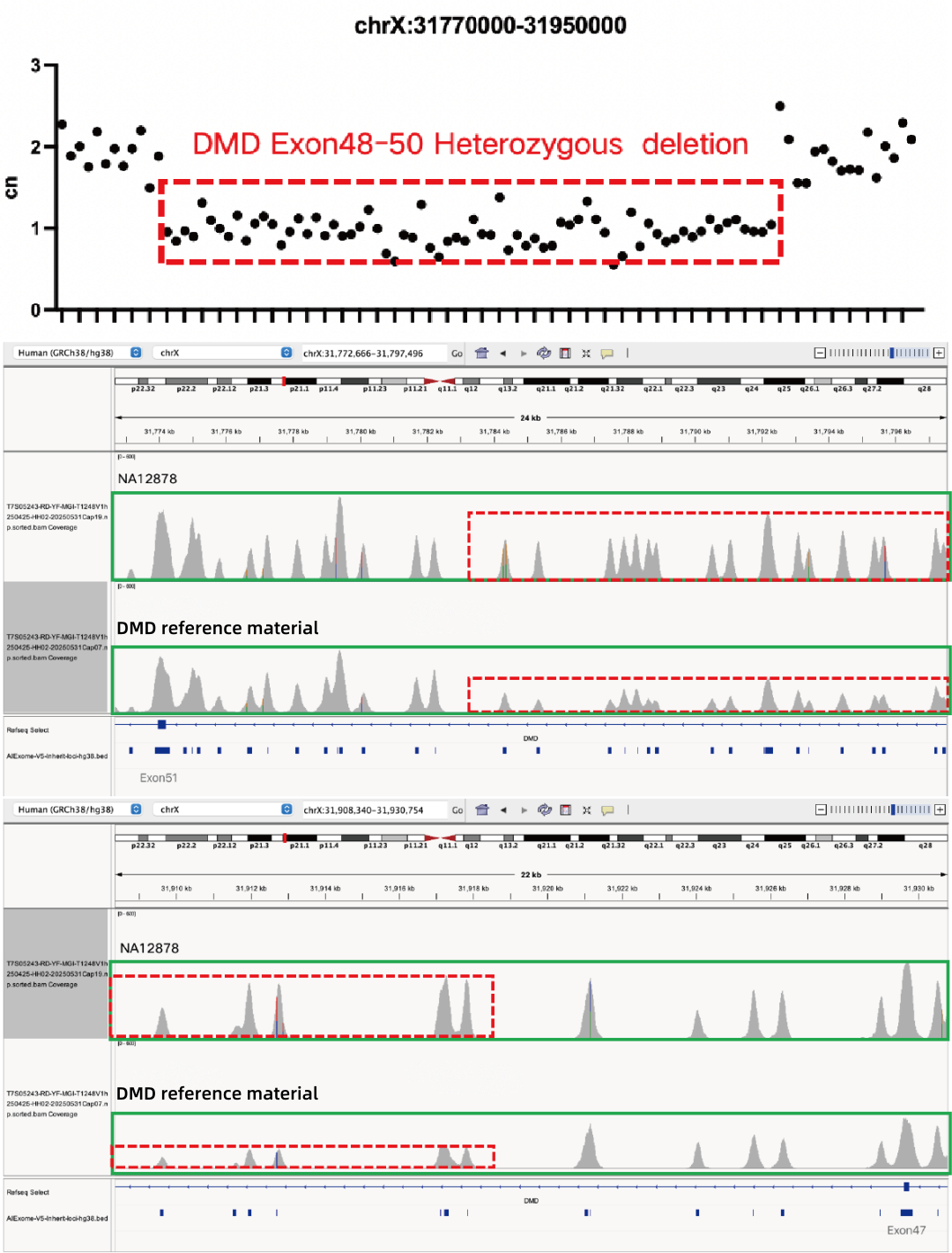
Evaluation of CNV Positive Control Detection Results
Using gDNA standard samples with an input of 100 ng, libraries were constructed with the IGT® Enzyme Plus Library Prep Kit V3, captured by AIExome® V5-Inherit, and sequenced by MGI PE150. The method successfully detected multiple common deletion types of thalassemia, and single-exon level CNVs in the DMD gene.
Second Layer of Safeguard: High-density probes coverage for detection of Large Fragment Deletions
AIExome® V5-Inherit employs 10Kb high-density probe design for key CNV regions in multiple databases such as Decipher and ClinGen CNV, covering CNV regions of critical concern in various genetic disease detections—including high-risk pathogenic regions like 22q11.2 deletion, 1p36 deletion, and 17q12 deletion.
1p36 deletion syndrome, one of the most common chromosomal terminal deletion syndromes, involves the distal region of the short arm of chromosome 1. Common manifestations include intellectual disability, epilepsy, cardiac malformations, craniofacial abnormalities, etc.
Probe coverage status in the 1p36 region
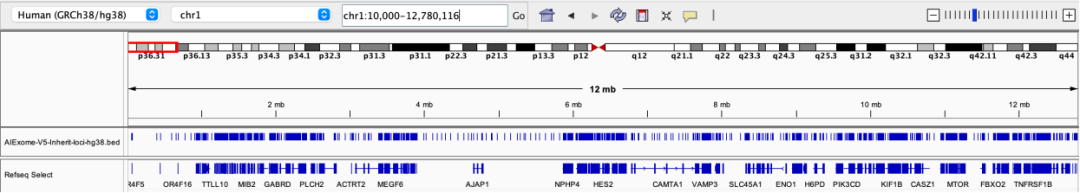
17q12 deletion syndrome is a chromosomal microdeletion syndrome involving key genes such as HNF1B, with common manifestations including renal cystic dysplasia, diabetes, intellectual disability, epilepsy, and autism spectrum disorder.
Probe coverage status in the 17q12 region
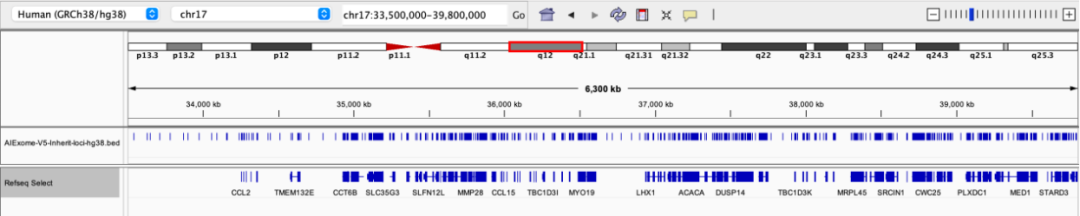
22q11.2 deletion syndrome is the most common microdeletion syndrome, involving key genes such as TBX1. Clinical manifestations include congenital heart disease, immunodeficiency, cleft palate, facial dysmorphism, intellectual disability, and mental illnesses (such as schizophrenia), featuring high phenotypic diversity.
Probe coverage status in the 22q11 region
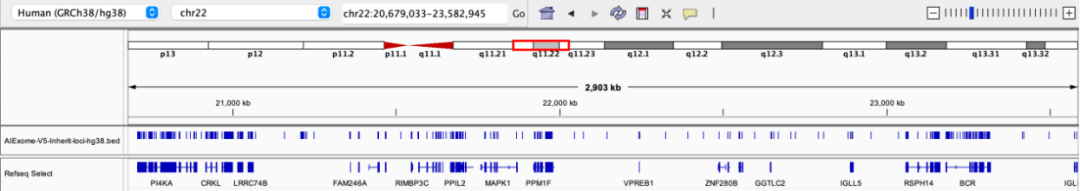
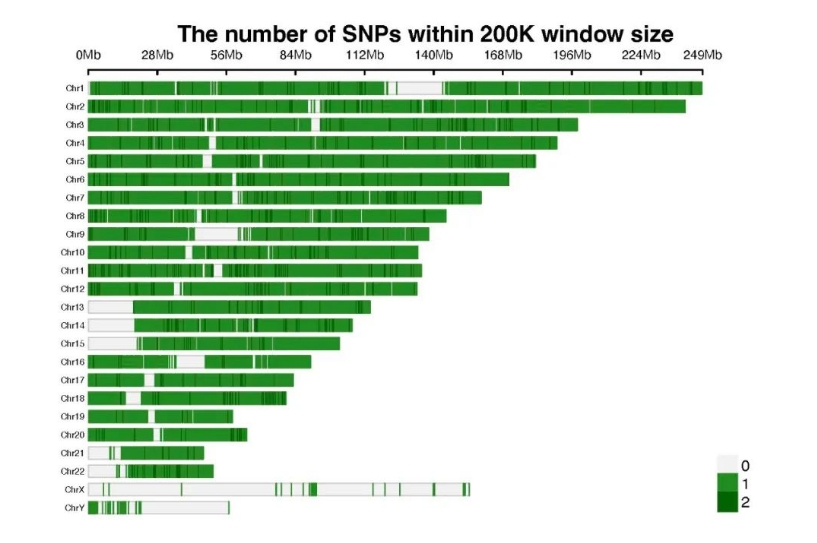
Third Layer of Safeguard: Whole-genome CNV backbone with 200 kb spacing, also available in 50/100 kb options
The Inherit Whole Exome default configuration includes a whole-genome CNV skeleton with 200 kb spacing. Probes are evenly distributed to cover intergenic regions and structurally complex areas, suitable for whole-genome CNV profiling, inherit disease screening, and population studies. Compared with high-density designs, this approach requires smaller data volume and simpler analysis. When combined with high-density probes in key regions, it enables a hierarchical detection strategy to enhance overall detection coverage and efficiency.
Highlight 2: High Coverage of ClinVar Database to Facilitate Clinical Interpretation
AIExome® V5-Inherit comprehensively covers mutation sites of significant clinical significance in the ClinVar database, primarily including variants related to "Pathogenic (P)", "Likely pathogenic (LP)", and "Drug response (DR)". These sites are typically associated with specific inherit diseases or tumor susceptibility, supported by clear literature evidence or consistent conclusions from multiple clinical laboratories. For example, frequent mutations in common disease genes such as BRCA1/2, CFTR, TP53, and HBB are widely used in clinical diagnosis and inherit counseling.
Probe Capture Test Data of AIExome V5-Inherit
Analysis of coverage and uniformity for Pathogenic/Likely Pathogenic (P/LP) mutation sites in the ClinVar database shows:
-Tested coverage rate reaches 99.92%
-30% coverage rate achieves 99.65%
-50% xMeandepth coverage rate achieves 93.13%
These data demonstrate more accurate and reliable mutation analysis.

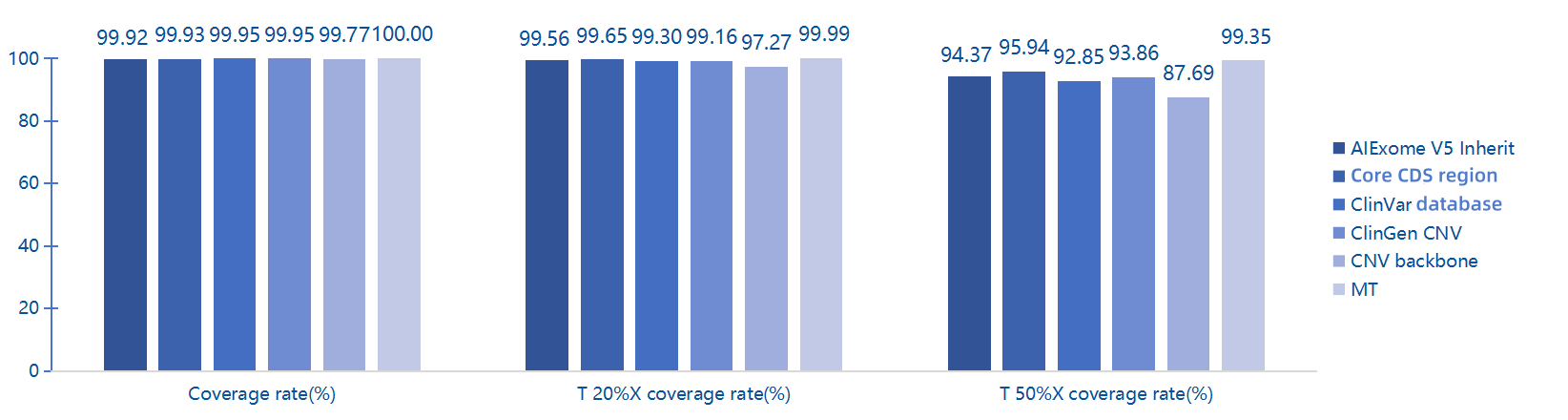
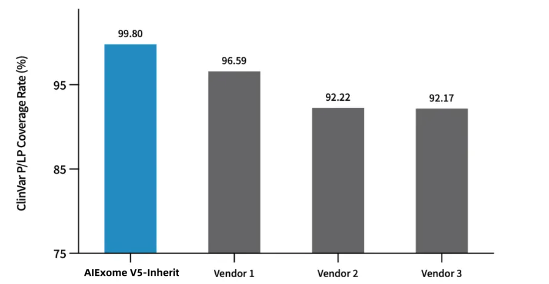
Coverage Statistics of Pathogenic and Likely Pathogenic Sites in ClinVar Database by Multiple Whole Exome Capture Products
AIExome V5-Inherit covers 99.8% of Pathogenic/Likely Pathogenic (P/LP) mutation sites in the ClinVar database.
Highlight 3: High Mutation Detection Accuracy for More Trustworthy Results
In inherit disease diagnosis, high detection accuracy effectively reduces the risks of misdiagnosis and missed diagnosis, ensuring the precise identification of key pathogenic variants and enhancing clinicians' confidence in clinical decision-making. Balancing high precision and recall helps comprehensively cover inherit variations, improving diagnostic reliability, which is crucial for enhancing the success rates of prenatal screening, personalized medicine, and rare disease diagnosis.
The detection performance of AIExome® V5-Inherit was evaluated using the NA12878 standard cell line, demonstrating excellent performance in SNP and Indel detection:
· SNP detection precision reached 99.74%, recall 99.71%, and F1-Score exceeded 99.72%;
· Indel detection F1-Score was also above 96%.
The product showed outstanding capability in identifying clinically relevant mutation sites in databases like ClinVar, reflecting its high reliability and accuracy in both clinical and research applications.
| SNP |
| Repeat 1 | Repeat 2 | Repeat 3 |
| NA12878 sites | 32825 |
| Sites detected | 32842 | 32803 | 32833 |
| Concordance of sites | 32740 | 32708 | 32738 |
| Recall(%) | 99.74 | 99.64 | 99.73 |
| Precision(%) | 99.69 | 99.71 | 99.71 |
| F1 Score(%) | 99.72 | 99.68 | 99.72 |
| InDel |
| Repeat 1 | Repeat 2 | Repeat 3 |
| NA12878 sites | 677 |
| Sites detected | 728 | 710 | 710 |
| Concordance of sites | 668 | 663 | 666 |
| Recall(%) | 98.67 | 97.93 | 98.38 |
| Precision(%) | 91.76 | 93.38 | 93.8 |
| F1 Score(%) | 95.09 | 95.6 | 96.03 |
*Variation data of the NA12878 standard sample and its high-confidence regions were selected, and the intersection with the target regions of AIExome® V5-Inherit was calculated. Within this intersection area, the reference dataset included 32,825 SNPs and 677 InDels. Based on this region, variation information from three sets of NA12878 test results was extracted for subsequent sensitivity and precision evaluation.
· Recall = TP/(TP + FN);
· Precision = TP/(TP + FP);
· F1-score = 2×Precision×Recall/(Precision+Recall).
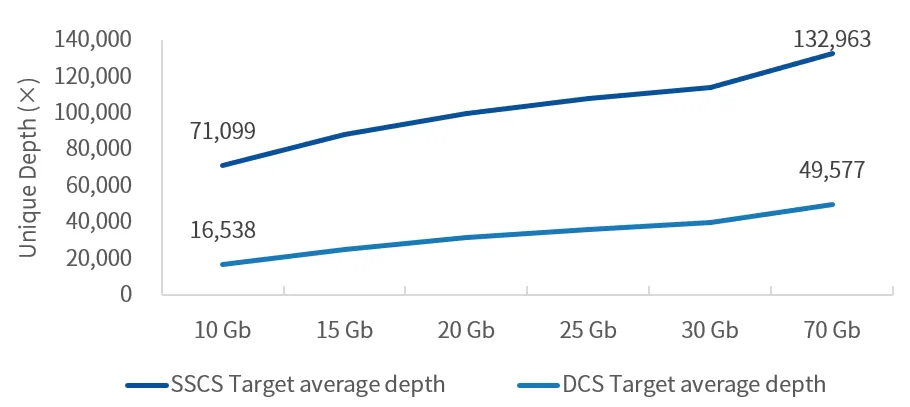
Highlight 4: Excellent Data Metrics with High Mitochondrial Sequencing Depth
While ensuring capture performance far exceeding the industry average, AIExome® V5-Inherit incorporates full-length mitochondrial probes via spike-in technology, enabling high-depth sequencing of mitochondrial DNA (mtDNA). This allows mutation detection for all 37 mitochondrial genes, covering point mutations, insertions/deletions, and heteroplasmy—suitable for screening and diagnosis of mitochondrial-related disorders such as hereditary muscular dystrophy, hereditary optic neuropathy, mitochondrial encephalomyopathy, and deafness.
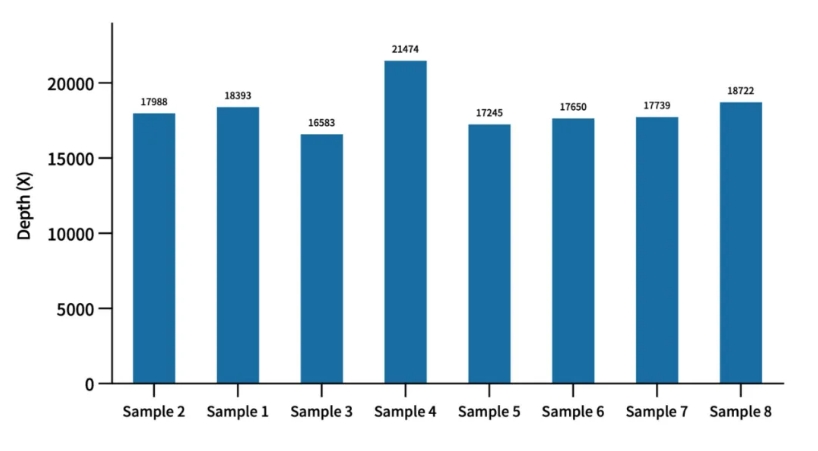
Mitochondrial Depth Evaluation of AIExome V5-Inherit Test Data
Using cell line quality control samples for enzymatic digestion library construction with a 750 ng library input and overnight hybridization, when the average depth of AIExome® V5-Inherit target regions reaches 100X, the average mitochondrial depth stably exceeds 10,000X.
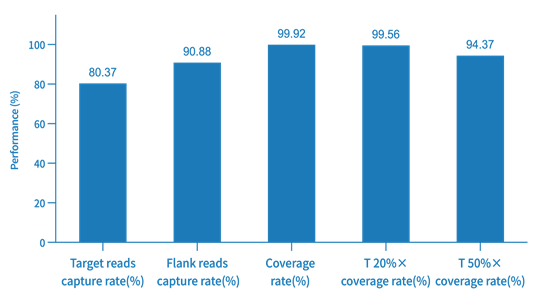
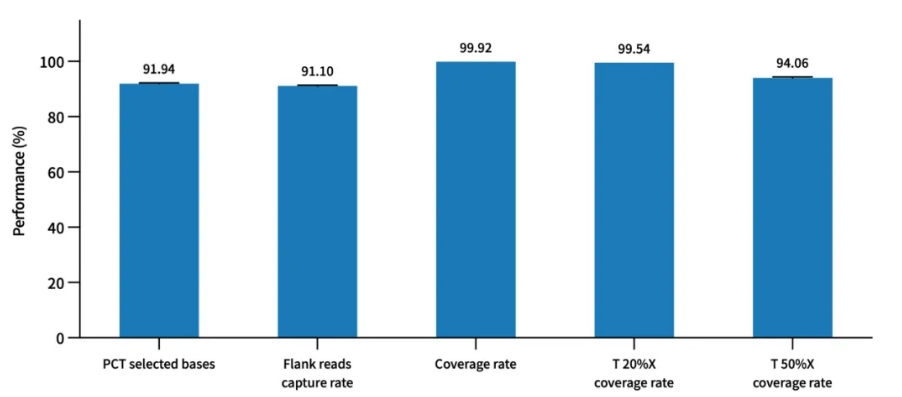
Probe Capture Performance of AIExome V5-Inherit with TargetSeq one v3.0 Overnight Hybridization
Enzymatic digestion library construction was performed using a pooled human standard sample (Promega, cat.G3041) with a 750 ng library input and overnight hybridization, followed by Illumina Novaseq 6000 PE150 sequencing.
Supports Multiple Plus Solutions: Flexible and Convenient Customization of Intellectual Property-Exclusive Whole Exome Products
· Spike-in with Numerous Predefined Product Probe Packs:Carefully designed for direct mixing and use, ensuring convenience and speed.
· 680K In-House Probe Synthesis Platform:Enables flexible and rapid response to requirements for adding/encrypting regions.
· 11-Year Expertise in Probe Design, Synthesis, and Reagent Development:Delivers superior and reliable product services.

 CN
CN